Figure 1.
Characterization of Open-Loop Transcriptional Sensors
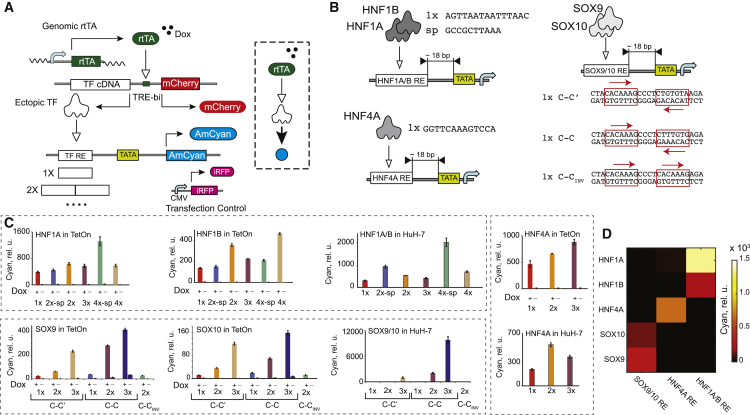
(A) Schematics of a sensor screening assay in HEK Tet-On cells. TATA indicates a core minimal promoter. DNA constructs and protein products are shown. Pointed arrow indicates transactivation.
(B) Schematics of the HNF1A/B, HNF4A, and SOX9/10 sensors. The sequences of 1× REs and a spacer (sp) are shown. Only top strands are shown for HNF1A/B and HNF4A.
(C) TF sensor population-averaged responses in HEK Tet-On cells in the presence and absence of Dox, as indicated, and in the HuH-7 cell line. Each bar represents mean ± SD of biological triplicates.
(D) Mutual crosstalk between the TFs (y axis) and sensors (x axis). The color code indicates averaged AmCyan intensity.
rtTA, reverse tetracycline-responsive transcriptional activator; CMV, cytomegalovirus promoter; TetOn, HEK293 Tet-On cells.