Abstract
We identified SA1684 as a Staphylococcus aureus virulence gene using a silkworm infection model. The SA1684 gene product carried the DUF402 domain, which is found in RNA-binding proteins, and had amino acid sequence similarity with a nucleoside diphosphatase, Streptomyces coelicolor SC4828 protein. The SA1684-deletion mutant exhibited drastically decreased virulence, in which the LD50 against silkworms was more than 10 times that of the parent strain. The SA1684-deletion mutant also exhibited decreased exotoxin production and colony-spreading ability. Purified SA1684 protein had Mn2+- or Co2+-dependent hydrolyzing activity against nucleoside diphosphates. Alanine substitutions of Tyr-88, Asp-106, and Asp-123/Glu-124, which are conserved between SA1684 and SC4828, diminished the nucleoside diphosphatase activity. Introduction of the wild-type SA1684 gene restored the hemolysin production of the SA1684-deletion mutant, whereas none of the alanine-substituted SA1684 mutant genes restored the hemolysin production. RNA sequence analysis revealed that SA1684 is required for the expression of the virulence regulatory genes agr, sarZ, and sarX, as well as metabolic genes involved in glycolysis and fermentation pathways. These findings suggest that the novel nucleoside diphosphatase SA1684 links metabolic pathways and virulence gene expression and plays an important role in S. aureus virulence.
Keywords: nucleoside/nucleotide metabolism, silkworm, Staphylococcus aureus (S. aureus), toxin, virulence factor, Staphylococcus aureus
Introduction
Staphylococcus aureus is a human pathogen that causes various diseases, including impetigo, meningitis, pneumonia, and sepsis. Methicillin-resistant S. aureus (MRSA)2 has been associated with serious clinical problems since the 1960s. The recent emergence of a new type of MRSA, called community-acquired MRSA, has become an especially urgent clinical concern. Only a few drugs, such as vancomycin, are available for treating MRSA diseases and novel pharmacotherapies are in high demand. S. aureus produces a wide array of virulence factors, including superantigens that interfere with host immune responses, cell wall proteins that facilitate bacterial adherence to host tissues, and extracellular toxins that damage host cells. Expression of these virulence factors is regulated by various factors, including agr, arlRS, and saeRS (1–3). Further identification of S. aureus virulence factors and their regulatory networks is important for establishing effective therapeutic strategies.
Recent studies suggest that nucleotide metabolism has an important role in S. aureus virulence gene expression. Mutations in the thyA gene encoding thymidylate synthase lead to growth defects, increased antibiotic resistance, and decreased expression of agr, a master regulator of S. aureus virulence genes, which are phenotypes of small colony variants (4–6). Knock-out of the thyA gene attenuates S. aureus virulence in mice and Caenorhabditis elegans (6). CodY is a transcription factor that binds GTP and regulates the transcription of S. aureus virulence genes (7). Two nucleotide-signaling molecules, (p)ppGpp and cyclic-di-GMP, also have roles in S. aureus virulence. The amount of (p)ppGpp increases when bacteria are starved and regulates the expression of various metabolic enzymes and virulence factors (8, 9). Cyclic di-GMP regulates biofilm formation and the expression of virulence factors (10, 11). The factors that link nucleotide metabolism and virulence gene expression, however, remain unclear.
We previously established an S. aureus infection model using silkworms, larvae of Bombyx mori, a lepidopteran insect species (12–14). Using this model, we revealed that virulence regulatory factors such as agr, arlRS, and saeRS are required for virulence of S. aureus against silkworms, suggesting that the silkworm model is useful for evaluating S. aureus virulence (15). We also identified virulence factors of S. aureus from hypothetical genes that are widely conserved in bacteria (16, 17), including a phosphodiesterase against RNA (18), an RNA-binding protein (19, 20), a regulatory factor for the expression of thyA (21), and rRNA methyltransferases (22, 23). These findings suggest that factors interacting with nucleic acid molecules play important functions in S. aureus virulence expression. In this study, we found that a factor with similarity to the S. coelicolor nucleoside diphosphatase, whose crystal structure was recently deposited in the Protein Data Bank (code 3EXM), is prominently involved in S. aureus virulence.
Results
Identification of S. aureus Virulence Factors by Evaluation of the S. aureus Killing Ability against Silkworms
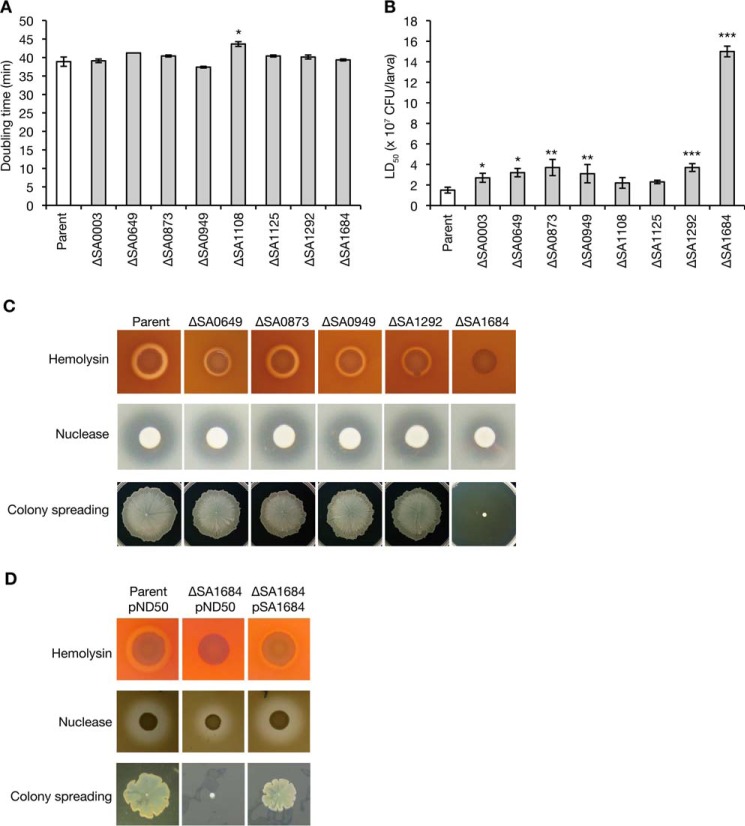
We selected eight genes from the S. aureus “conserved hypothetical genes” that encode proteins predicted to interact with nucleic acid molecules using an in silico database, pfam, and RefSeq (www.ncbi.nlm.nih.gov). S. aureus deletion mutants of these eight genes were constructed by double homologous recombination (24). The doubling time of seven of the gene-deletion mutants was indistinguishable from that of the parent strain. In contrast, the doubling time of the remaining deletion mutant, SA1108, was 1.12-fold longer than that of the parent strain (Fig. 1A). We next examined whether these gene-deletion mutants had attenuated killing abilities against silkworms by determining the LD50 values. The LD50 values of the gene-deletion mutants of SA0649, SA0873, SA0949, and SA1292 were more than 2-fold that of the parent strain (Fig. 1B). The LD50 value of the SA1684-deletion mutant was 10 times that of the parent strain (Fig. 1B). These findings suggest that SA0649, SA0873, SA0949, and SA1292 contribute to S. aureus virulence and that SA1684 plays a critical role in S. aureus virulence in silkworms.
FIGURE 1.
Evaluation of the silkworm killing ability, exotoxin production, and colony-spreading ability of the deletion mutants of novel virulence factors. A, overnight cultures of the S. aureus parent strain and gene-deletion mutants were inoculated to a 100-fold amount of fresh tryptic soy broth and cultured at 27 °C. Doubling time was calculated by the growth curves. Data are presented as means ± S.E. from two independent experiments. Asterisk indicates a Student's t test p value against the parent strain of less than 0.05. B, overnight cultures of S. aureus gene-deletion mutants were 2-fold serially diluted and injected into the silkworm hemolymph (n = 5), and survival was determined at 24 h after the injection. The colony-forming unit values that cause 50% of the silkworms to die (LD50) were determined from the survival curves. Data shown are means ± S.E. from independent measurement of LD50 (n = 14 for parent strain, n = 5 for gene-deletion mutants). Asterisks indicate the Student's t test p value against the wild-type strain (*, p < 0.05; **, p < 0.01; *** p < 0.001). The SA0003-deletion mutant had a significantly increased LD50 value, but it was less than 2-fold that of the parent strain. C, exotoxin production and colony-spreading ability of the parent, ΔSA0649, ΔSA0873, ΔSA0949, ΔSA1292, and ΔSA1684 strains were evaluated. Hemolysin and nuclease production was examined by spotting overnight cultures onto sheep blood agar plates or DNA agar plates (hemolysin, upper panel; nuclease, middle panel). Colony spreading was examined by culturing S. aureus cells on soft agar plates (lower panel). Data are representative of three independent experiments. D, exotoxin production and colony spreading of the parent strain transformed with empty vector (pND50) and ΔSA1684 strain transformed with empty vector or pSA1684 were evaluated.
The functional domains of these five proteins were examined and are summarized in Table 1. The SA0649 product carries DUF296, which is found in DNA-binding proteins carrying an AT hook motif. The SA0873 product carries a domain that is found in 2′,5′-RNA ligase and shares 36% amino acid sequence similarity with the Bacillus subtilis YjcG protein (25, 26). The SA0949 product carries an HTH_3 domain, which is found in bacterial proteins maintaining plasmid copy number, bacterial DNA methylase, and bacterial transcription factor. The SA1292 product carries a nucleotide pyrophosphohydrolase domain found in MazG that hydrolyzes nucleoside triphosphate into nucleoside monophosphate and pyrophosphate. Escherichia coli MazG interacts with GTPase Era, which has multicellular functions (27) and is required for survival in an amino acid-starved condition (28, 29). The SA1684 product carries DUF402 domain, which is found in RNase E or RNase G.
TABLE 1.
Domains of novel virulence factors
Domains of novel virulence factors identified in this study were searched by the in silico programs pfam and Ref Seq. ORF No. is ID in the S. aureus N315 genome database. In the search using the pfam program, domains with an e-value less than e-4 are presented.
| ORF no. | Length in amino acids | Region | pfam |
Ref. sequence protein function | ||
|---|---|---|---|---|---|---|
| e-value | pfamID | Domain Function | ||||
| SA0649 | 140 | 9-119 | 1.4 × e−18 | DUF296 | Domain of unknown function | Predicted DNA-binding protein |
| SA0873 | 169 | 9-3 | 6.9 × e−9 | LigT_PEase | 2′,5′-RNA ligase | |
| 10-156 | 1.4 × e−19 | 2_5_RNA_ligase2 | ||||
| SA0949 | 179 | 4-65 | 4.8 × e−17 | HTH_19 | Helix-turn-helix domain | Uncharacterized |
| 4-60 | 1.6 × e−8 | HTH_31 | ||||
| 7-61 | 1.4 × e−14 | HTH_3 | Helix-turn-helix | |||
| 106-173 | 8.2 × e−10 | Cupin_2 | Cupin domain | |||
| 10-37 | 2.5 × e−5 | Sigma70_r4 | Sigma-70, region 4 | |||
| SA1292 | 180 | 23-102 | 7.4 × e−22 | MazG | MazG nucleotide pyrophosphohydrolase domain | Predicted pyrophosphatase |
| 2-92 | 8.9 × e−5 | dUTPase_2 | dUTPase | |||
| SA1684 | 180 | 63-127 | 7.5 × e−18 | DUF402 | Protein of unknown function | Associate with RNase E and RNase G |
Contribution of Novel Virulence Factors to Extracellular Toxin Production and Colony Spreading
To reveal the functions of novel virulence genes, we examined the amount of S. aureus extracellular toxins, including hemolysin and nuclease, and colony spreading, which is the expansion of the S. aureus on soft agar surfaces (30). The SA1684-deletion mutant had decreased hemolysin production, nuclease production, and colony-spreading ability compared with the parent strain (Fig. 1C). The SA0649-, SA0949-, and SA1292-deletion mutants exhibited slightly decreased hemolysin production, whereas the nuclease production and colony spreading did not differ from that of the parent strain (Fig. 1C). The SA0873-deletion mutant was indistinguishable from the parent strain with regard to hemolysin and nuclease production and colony spreading. Introduction of SA1684 to the SA1684-deletion mutant restored the hemolysin production, nuclease production, and the colony-spreading activity (Fig. 1D). These results suggest that SA0649, SA0949, and SA1292 contribute only slightly to S. aureus hemolysin production and that SA1684 is required for hemolysin production, nuclease production, and colony spreading.
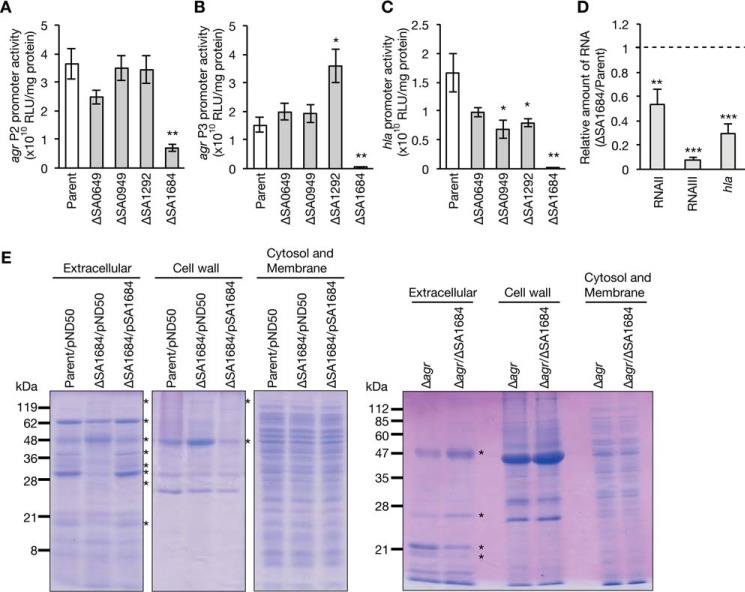
In S. aureus, the transcription of hemolysin genes is positively regulated by the agr locus (31). The P2 transcript of the agr locus encodes AgrD, AgrB, AgrC, and AgrA, which coordinately function in quorum sensing. The P3 transcript of the agr locus functions as a regulatory RNA, called RNAIII, and regulates the expression of various virulence factors (1). To examine whether the decreased hemolysin production of the SA0649-, SA0949-, SA1292-, and SA1684-deletion mutants was due to the decreased expression of agr, we measured the promoter activities of agr and hla encoding α-hemolysin. In the SA1684-deletion mutant, the activities of agr P2 and agr P3 were decreased to less than 20% that of parent strain (Fig. 2, A and B). In addition, in the SA1684-deletion mutant, the activity of the hla promoter was decreased to less than 10% that of the parent strain (Fig. 2C). Furthermore, the amounts of the P2 and P3 transcripts (RNAII and RNAIII) of the agr locus as well as the amount of hla mRNA were decreased in the SA1684-deletion mutant compared with the parent strain (Fig. 2D). In the SA0949- and SA1292-deletion mutants, the activities of agr P2 and agr P3 were not decreased, but the activity of the hla promoter was slightly decreased (Fig. 2, A–C). In the SA0649-deletion mutant, the activities of agr P2, agr P3, and the hla promoter were not decreased (Fig. 2, A–C). These results suggest that SA1684 is required for expression of the agr locus and the hla gene, whereas SA0649, SA0949, and SA1292 are not required for agr expression.
FIGURE 2.
Expression of the agr locus and the hla gene in the deletion mutants of novel virulence genes. A–C, S. aureus parent, ΔSA0649, ΔSA0873, ΔSA0949, ΔSA1292, and ΔSA1684 strains were transformed with reporter fusion plasmids carrying agr P2 (A), agr P3 (B), or hla promoter (C), and cultured to A600 =1.0. Luciferase activities were measured. Data are presented as means ± S.E. from three independent experiments. Asterisk represents the Student's t test p value between the gene-deletion mutant and the parent strain (*, p < 0.05; **, p < 0.01). D, S. aureus parent strain and ΔSA1684 strain were cultured to A600 = 1, and total RNA was extracted. Amounts of RNAII, RNAIII, hla mRNA, and 16S rRNA were measured by quantitative reverse transcription-PCR. Data were normalized with the amount of 16S rRNA and are presented as relative value to that of the parent strain. Data are presented as means ± S.E. from three independent experiments. Asterisk represents the Student's t test p value between the parent strain and ΔSA1684; (**, p < 0.01; *** p < 0.001). E, extracellular proteins, cell wall proteins, and cytosol/membrane proteins of S. aureus overnight cultures were electrophoresed in SDS-polyacrylamide gel, and stained with Coomassie Brilliant Blue. Left three gel panels show the result of the parent strain transformed with empty vector, ΔSA1684 strain transformed with empty vector, or pSA1684. Right gel panel shows the result of Δagr strain and Δagr/ΔSA1684 strain. Asterisks indicate the protein band whose intensity differs between Parent/pND50 and ΔSA1684/pND50 or between Δagr and Δagr/ΔSA1684.
Based on the above results suggesting that SA1684 has a crucial role in S. aureus virulence, we further investigated the function of SA1684. In S. aureus, the agr locus regulates the expression of various extracellular proteins and cell wall proteins. We examined whether SA1684 contributes to the expression of extracellular proteins, cell wall proteins, and cytosol/membrane proteins. In the SA1684-deletion mutant, several proteins in the extracellular fraction and cell wall fraction were differentially expressed compared with the parent strain (Fig. 2E). In contrast, there were no differences between the parent strain and the SA1684-deletion mutant in the cytosol/membrane fraction (Fig. 2E). The differential expression of the extracellular proteins and cell wall proteins in the SA1684-deletion mutant was cancelled by introducing the intact SA1684 gene (Fig. 2E). These results suggest that SA1684 contributes to the expression of extracellular proteins and cell wall proteins. In the agr-deletion background, the deletion of SA1684 affected the expression of several extracellular proteins (Fig. 2E). The results suggest that SA1684 contributes to the expression of several extracellular proteins in an agr-independent manner.
SA1684 Protein Has Mn2+-dependent Hydrolyzing Activity against NDPs
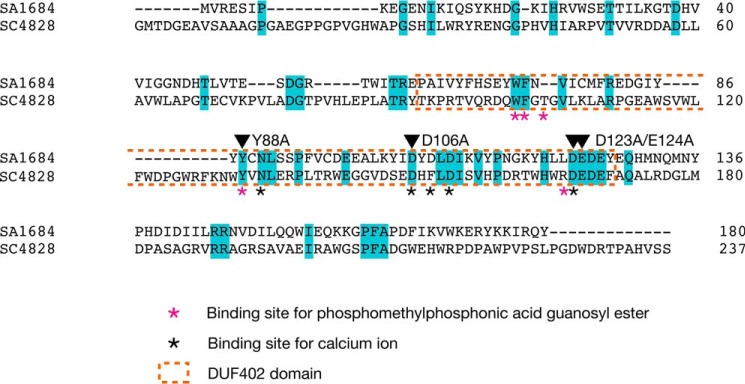
SA1684 protein has the DUF402 domain that is found in RNA-binding proteins such as RNase E or RNase G and exhibited 22% amino acid identity with S. coelicolor SC4828 protein, for which a cocrystal structure with phosphomethylphosphonic acid guanosyl ester, a non-hydrolyzing GDP analog, was recently deposited in the Protein Data Bank (3EXM) (Fig. 3). We examined whether the SA1684 protein has nucleoside diphosphatase activity by purifying a recombinant C-terminal His-tagged SA1684 protein from E. coli that was transformed with the SA1684-overproducing plasmid. SDS-PAGE analysis revealed that the recombinant SA1684 protein was purified to more than 90% purity, and the molecular mass was 28 kDa (Fig. 4A). Recombinant SA1684 protein hydrolyzed UDP to UMP in the presence of Mn2+ ions, whereas the protein purified from E. coli transformed with empty plasmid did not hydrolyze UDP (Fig. 4B). SA1684 protein hydrolyzed UDP in both a dose-dependent manner (Fig. 4C) and a time-dependent manner (Fig. 4D). The results suggest that SA1684 protein has hydrolyzing activity against UDP. UDP hydrolysis by SA1684 was observed in the presence of Mn2+ or Co2+ ions but not in the presence of Cu2+, Ni2+, Zn2+, Mg2+, or Ca2+ ions at a 0.5 mm concentration (Fig. 4E). UDP hydrolysis occurred with Mn2+ ions at concentrations of 0.5–12.5 mm and slightly occurred with Mg2+ ions at a 12.5 mm concentration (Fig. 4F).
FIGURE 3.
Amino acid sequence similarity between SA1684 and SC4828 proteins. Amino acid sequences of SA1684 and SC4828 were aligned using ClustalW. Identical amino acids are colored cyan. Asterisks indicate the amino acids important for the interaction with Ca2+ ions or phosphomethylphosphonic acid guanosyl ester in the crystal structure of SC4828. Orange-dotted region indicates the DUF402 domain. Arrowheads indicate the amino acid residues substituted with alanine in this study.
FIGURE 4.
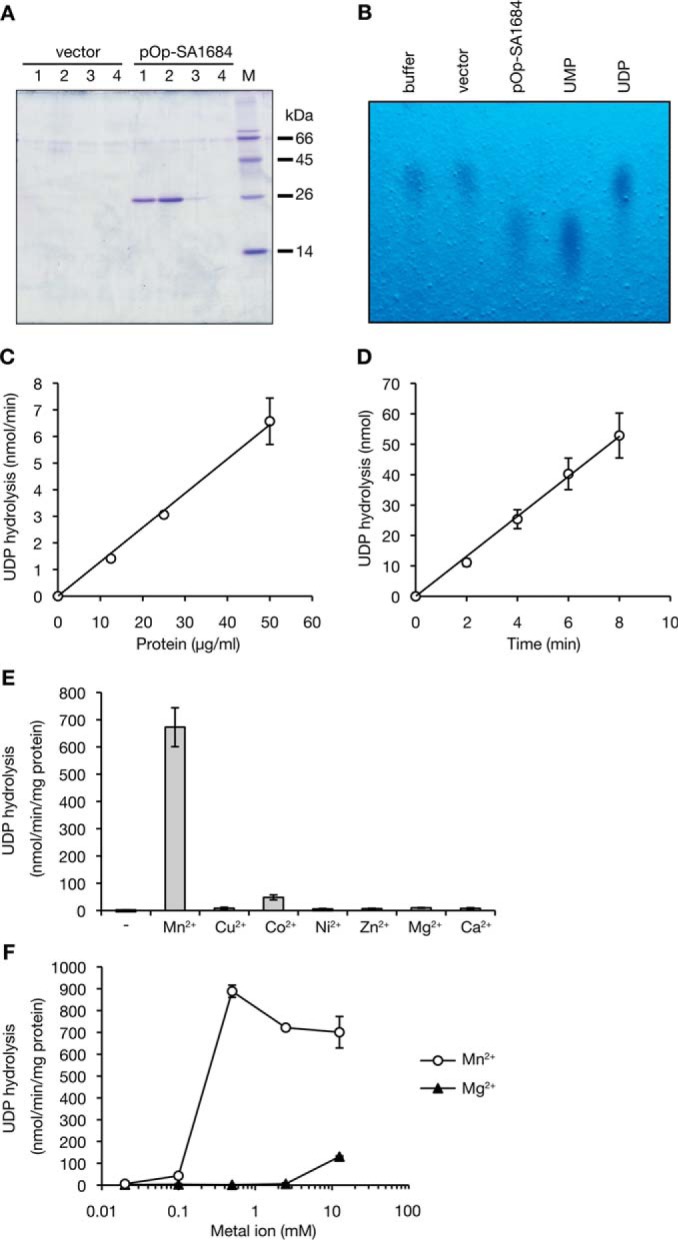
SA1684 protein has hydrolytic activity against UDP in the presence of Mn2+ ions. A, protein was purified using a nickel affinity column from an E. coli strain transformed with empty vector or pOp-SA1684. Proteins were electrophoresed in 15% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. Lanes 1–4 indicates fraction numbers eluted with imidazole. M indicates marker proteins. B, purified protein from E. coli transformed with empty vector (2.7 μg/ml) or with pOp-SA1684 (50 μg/ml) was reacted with 1 mm UDP in the presence of Mn2+ for 240 min at 26 °C. The reaction product was analyzed by TLC. UDP or UMP was visualized by UV light. Buffer indicates the sample without protein addition. C, dose dependence of SA1684 on UDP hydrolysis was examined. Different amounts of SA1684 protein were reacted with 1 mm UDP in the presence of Mn2+ ions for 2, 4, 6, or 8 min at 26 °C. Phosphate released per min was measured. Data are presented as means ± standard errors from three independent experiments. D, time dependence of UDP hydrolysis by SA1684 was examined. SA1684 protein (50 μg/ml) was reacted with 1 mm UDP in the presence of Mn2+ ions for 2, 4, 6, or 8 min at 26 °C. Phosphate released was measured. Data are presented as means ± S.E. from three independent experiments. E, metal dependence of UDP hydrolysis by SA1684 was examined. SA1684 protein (50 μg/ml) was reacted with 1 mm UDP in the presence of 0.5 mm various metal ions for 8 min at 22 °C. Released phosphates per min were measured. Data are presented as means ± S.E. from triplicate assays. F, dose response of Mn2+ or Mg2+ ions on UDP hydrolysis by SA1684 was examined. SA1684 protein (50 μg/ml) was reacted with 1 mm UDP in the presence of different concentrations of Mn2+ or Mg2+ ions (0.02, 0.1, 0.5, 2.5, or 12.5 mm). Phosphate released per min was measured. Data are presented as means ± S.E. from triplicate assays.
SA1684 protein hydrolyzed five NDPs (ADP, GDP, UDP, CDP, and IDP) and four dNDPs (dADP, dGDP, TDP, and dCDP) but did not hydrolyze nucleoside monophosphates or nucleoside triphosphates (Fig. 5A). We determined the Km and Vmax values for SA1684 protein hydrolysis of ADP, GDP, UDP, CDP, and TDP. The Km values for these nucleotides ranged between 96 and 300 μm, and the Vmax values ranged between 0.99 and 3.8 μmol/min/mg protein (Fig. 5B and Table 2). Hydrolysis of UDP and GDP by SA1684 protein was inhibited by high concentrations of the substrate (Fig. 5B).
FIGURE 5.
Substrate specificity of SA1684. A, SA1684 protein (50 μg/ml) was reacted with 1 mm nucleoside monophosphate (AMP, GMP, UMP, and CMP), deoxynucleoside monophosphate (TMP), nucleoside diphosphate (ADP, GDP, UDP, CDP, and IDP), deoxynucleoside diphosphate (dADP, dGDP, TDP, and dCDP), nucleoside triphosphate (ATP, GTP, UTP, and CTP), or deoxynucleoside triphosphate (TTP) in the presence of Mn2+ ions for 8 min at 26 °C. Phosphates released per min were measured. Data are presented as means ± S.E. from three independent experiments. B, 2-fold serial concentrations (62.5, 125, 250, 500, and 1000 μm) of ADP, UDP, CDP, GDP, and TDP were reacted with SA1684 protein (12.5 μg/ml) in the presence of Mn2+ ions for 2, 4, or 6 min at 26 °C. Phosphates released per min were measured. Data are presented as means ± S.E. from three independent experiments and were fitted to Michaelis-Menten kinetics using nonlinear regression analysis. The data for UDP and GDP hydrolyzed at 1000 μm concentration could not be fitted to Michaelis-Menten kinetics or substrate inhibition kinetics. C, SA1684 protein (50 μg/ml) was reacted with 250 μm GDP in the absence or presence of 750 μm GDPβS or GTPγS for 8 min at 26 °C. Data are presented as means ± S.E. from triplicate assays.
TABLE 2.
Kinetic parameters of NDP hydrolysis by SA1684 protein
The kinetic parameters for GDP and UDP are based on the plots of lower substrate concentration (0–500 μm).
| ADP | GDP | CDP | UDP | TDP | |
|---|---|---|---|---|---|
| Km (μm) | 96.3 ± 34.1 | 102 ± 31 | 303 ± 54 | 221 ± 105 | 203 ± 59 |
| Vmax (μmol/min/mg protein) | 1.15 ± 0.11 | 0.985 ± 0.100 | 2.11 ± 0.15 | 3.84 ± 0.82 | 1.88 ± 0.19 |
| kcat (min) | 24.9 ± 2.4 | 21.4 ± 2.2 | 45.8 ± 3.3 | 83.3 ± 17.8 | 40.8 ± 4.2 |
To characterize the NDP hydrolysis by SA1684, we examined whether GDPβS or GTPγS, which are, respectively, non-hydrolyzable analogs of GDP or GTP, inhibits GDP hydrolysis by SA1684. Addition of a 3-fold molar amount of both analogs inhibited the GDP hydrolysis by SA1684 (Fig. 5C).
Amino Acid Residues Tyr-88, Asp-106, and Asp-123/Glu-124 of SA1684 Protein Are Required for NDP Hydrolysis and S. aureus Virulence
Crystal structure analysis of S. coelicolor SC4828 protein provided clues to the amino acid residues important for NDP hydrolysis (Protein Data Bank ID, 3EXM). The Tyr-88 and Asp-106 residues of the SA1684 protein correspond to the amino acid residues of SC4828 protein that are involved in binding to the substrate or calcium ions (Fig. 3). Asp-123 and Glu-124 form characteristic acidic amino acid residues (DEDE) that are conserved between SA1684 and SC4828 (Fig. 3). To examine whether Tyr-88, Asp-106, and Asp-123/Glu-124 are required for UDP hydrolysis by the SA1684 protein, we introduced alanine substitution mutations for these amino acids and purified the mutated SA1684 proteins as in the case of wild-type SA1684 protein (Fig. 6A). The Y88A, D106A, and D123A/E124A mutant SA1684 proteins diminished UDP hydrolysis activity (Fig. 6B). These results indicate that Tyr-88, Asp-106, and Asp-123/Glu-124 of SA1684 protein are required for the UDP hydrolysis activity.
FIGURE 6.
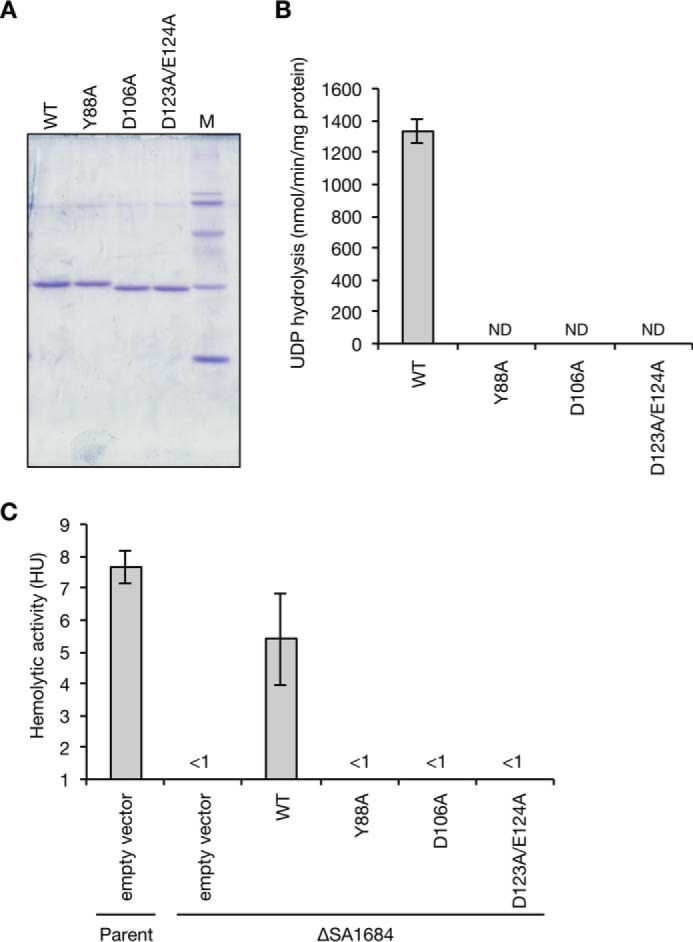
Effects of amino acid substitutions of SA1684 protein on the UDP hydrolysis and complementation activity against hemolysin production in the SA1684-deletion mutant. A, alanine-substituted SA1684 mutant proteins (Y88A, D106A, and D123A/E124A) were purified from E. coli-overproducing strains by the same method for wild-type SA1684 protein (WT). The purified proteins (1 μg) were electrophoresed in 15% SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue. B, UDP hydrolytic activities of mutant SA1684 proteins and wild-type SA1684 protein were measured at 26 °C in the presence of Mn2+ ions. Data are presented as means ± S.E. from three independent experiments. ND means not detected. C, S. aureus parent strain transformed with empty vector and SA1684-deletion mutant transformed with empty vector, wild-type SA1684 gene (WT), or mutant SA1684 genes encoding Y88A, D106A, D123A/E124A mutant proteins were cultured overnight. Hemolytic activity against sheep erythrocytes in the culture supernatant was measured. Data are presented as means ± S.E. from 3 to 5 independent experiments.
To determine whether Tyr-88, Asp-106, and Asp-123/Glu-124 of SA1684 protein are required for S. aureus virulence, we examined whether transformation with the mutant genes encoding the mutated SA1684 proteins restored the virulence of the SA1684-deletion mutant. Transformation with the wild-type SA1684 gene restored the hemolysin production of the SA1684-deletion mutant, whereas transformation of the mutated SA1684 genes encoding Y88A, D106A, D123A/E123A proteins did not restore the hemolysin production of the SA1684-deletion mutant (Fig. 6C). These results suggest that Tyr-88, Asp-106, and Asp-123/Glu-124 of SA1684 protein are required for S. aureus virulence.
Deletion of the SA1684 Gene Affects the Expression of Genes Involved in Glycolytic and Fermentation Pathways
To reveal the effects of SA1684 protein against S. aureus gene expression, we performed RNA sequence analysis using the S. aureus parent strain and the SA1684-deletion mutant, and we identified pathways affected by the SA1684 deletion based on gene enrichment analysis. In the SA1684-deletion mutant, a two-component system as well as several metabolic pathways, including those for glycolysis and pyrimidine metabolism, were affected (Table 3). We further identified individual genes whose expression was increased or decreased more than 2-fold in the SA1684-deletion mutant compared with the parent strain, and the effects were cancelled by introducing the SA1684 gene (supplemental Tables S1 and S2). Genetic manipulations of the SA1684 gene in the SA1684-deletion mutant and the SA1684-complemented strain were confirmed by the finding that the amount of SA1684 mRNA was considerably decreased in the SA1684-deletion mutant and restored in the SA1684-complemented strain (supplemental Table S1). Consistent with the results obtained by quantitative reverse transcription-PCR analysis (Fig. 2D) and gene enrichment analysis (Table 3), the RNA amounts of agrB, agrC, and hld (RNAIII), which encode a two-component system involved in virulence regulation, were decreased in the SA1684-deletion mutant compared with the parent strain, and the decreased expression was restored in the complemented strain (supplemental Table S1). The RNA levels of sarX and sarZ, which encode transcription factors that regulate the expression of virulence genes (17, 33), were decreased in the SA1684-deletion mutant compared with the parent strain (supplemental Table S1). In addition to these virulence regulatory factors, the RNA levels of pgk and tpiA, which encode enzymes of glycolysis, and adh1, ldh1, and ldh2, which encode enzymes of the fermentation pathway, were decreased in the SA1684-deletion mutant compared with the parent strain (supplemental Table S1). In contrast, the RNA amounts of purE and purK involved in inosine monophosphate synthesis and that of pfs, which encodes an enzyme to produce adenosine from 5′-methylthioadenosine or S-adenosylhomocysteine, were increased in the SA1684-deletion mutant compared with the parent strain (supplemental Table S2). These results suggest that deletion of the SA1684 gene alters the expression of various genes, such as genes involved in nucleotide metabolism, glycolysis, the fermentation pathway, and virulence regulation.
TABLE 3.
Gene enrichment analysis of differentially expressed genes in the SA1684-deletion mutant
FDR indicates false discovery rate.
| KEGG pathway | No. of genes in pathway | Normalized enrichment score | Normalized p value | FDR q-value |
|---|---|---|---|---|
| Down-regulated pathways in the SA1684 deletion mutant compared with the parent strain | ||||
| Oxidative phosphorylation | 22 | −2.23 | 0.000 | 0.000 |
| Pyrimidine metabolism | 46 | −1.84 | 0.000 | 0.011 |
| Two-component system | 63 | −1.82 | 0.000 | 0.009 |
| Alanine, aspartate and glutamate metabolism | 18 | −1.60 | 0.036 | 0.034 |
| Glycolysis/gluconeogenesis | 38 | −1.44 | 0.022 | 0.073 |
| Up-regulated pathways in the SA1684-deletion mutant compared with the parent strain | ||||
| Ribosome | 54 | 1.72 | 0.000 | 0.044 |
| Protein export | 16 | 1.55 | 0.016 | 0.185 |
| Folate biosynthesis | 18 | 1.54 | 0.029 | 0.139 |
| Fatty acid metabolism | 18 | 1.52 | 0.023 | 0.140 |
| ABC transporters | 99 | 1.46 | 0.007 | 0.207 |
Discussion
In this study, we searched for S. aureus virulence factors from hypothetical proteins interacting with nucleic acid molecules and identified a novel nucleoside diphosphatase necessary for S. aureus virulence. Amino acid-substituted mutant SA1684 proteins lost both their nucleoside diphosphatase activity and complementation activity against hemolysin production of the SA1684-deletion mutant, suggesting that the nucleoside diphosphatase activity of the SA1684 protein is required for S. aureus virulence. This study is the first to reveal the requirement of nucleoside diphosphatase in S. aureus virulence expression.
In the SA1684-deletion mutant, hemolysin production and colony-spreading activity were decreased. In addition, the promoter activities of the agr locus were decreased, and the amounts of agr transcripts (RNAII and RNAIII) were decreased in the SA1684-deletion mutant. Because the agr locus is required for hemolysin production and colony-spreading activity (34, 35), decreased expression of the agr locus is a plausible explanation for the decreased hemolysin production and the decreased colony-spreading activity of the SA1684-deletion mutant. Based on the finding that the nucleoside diphosphatase activity of SA1684 is required for S. aureus hemolysin production, we assume that alterations of the nucleotide amount by SA1684 protein is important for maintaining agr expression in S. aureus. Additional studies to measure the nucleotide pool in the SA1684-deletion mutant will be important to identify the nucleotide molecules important for the regulation of S. aureus virulence.
RNA sequence analysis revealed that deletion of SA1684 increases the expression of purE, purK, and pfs, which are involved in nucleotide metabolism. The purE and purK genes are involved in inosine monophosphate synthesis, which is a starting material for purine nucleotide synthesis. pfs encodes 5′-methylthioadenosine/S-adenosylhomocysteine nucleotidase to produce adenine from 5′-methylthioadenosine or S-adenosylhomocysteine, respectively. The increased expression of these nucleotide synthetic genes may be due to a compensatory reaction to the deficiency of nucleotides caused by the absence of nucleoside diphosphatase in the SA1684-deletion mutant. In addition to the nucleotide metabolic genes, expression of pgk, tpiA, adh1, ldh1, and ldh2, which are involved in glycolysis and fermentation pathways, was decreased in the SA1684-deletion mutant, suggesting that alterations of nucleotide metabolism lead to a decrease in the enzymes involved in glycolysis and fermentation pathways. Interestingly, in the deletion mutant of the cvfA gene encoding the RNA modification enzyme necessary for S. aureus virulence (16, 36), expression of adh1 and ldh2, which is involved in the fermentation pathway as well as expression of the virulence regulatory factors agr, sarX, and sarZ, is decreased compared with the parent strain (17, 37). Furthermore, in the hemB or menD mutant with the small colony variant phenotype of S. aureus, the expressions of pgk and tpi involved in glycolysis, ldh1 and pflB involved in the fermentation pathway, and the agr locus are decreased (38). In contrast, the deletion of agr does not decrease the expression of genes involved in the glycolysis and fermentation pathways (39–41). These findings suggest that the decreased gene expression involved in the glycolysis and fermentation pathways causes a decrease in agr expression. CodY regulates agr expression by binding to GTP (42, 43) or by responding to nutrient levels (44). Based on these previous findings and our present finding, we assume that CodY or other transcription factors recognize(s) alterations of the nucleotide pool or some metabolites in the glycolysis or fermentation pathways in the SA1684-deletion mutant, leading to the decreased agr transcription. Further investigation is required to reveal the molecular mechanisms by which alterations of the nucleotide pool lead to transcriptional regulation of the agr locus.
The LD50 value of the SA1684-deletion mutant against silkworms was more than 10-fold that of the parent strain. In contrast, agr deletion leads to a 3-fold increase in the LD50 value in the NCTC8325-4 strain, which expresses high amounts of agr (15). Thus, decreased virulence of the SA1684-deletion mutant against silkworms is not due only to decreased agr expression. Furthermore, deletion of SA1684 affected the expression of several extracellular proteins by an agr-independent pathway. The molecular mechanism underlying the nucleotide metabolism by SA1684 protein that leads to the agr-independent virulence expression remains to be clarified.
Biochemical characterization of SA1684 protein revealed that SA1684 shows hydrolytic activity to NDPs, but not to NMPs and NTPs. Because the GDP hydrolysis by SA1684 protein was inhibited by GDPβS as well as GTPγS, SA1684 might capture NTP but not hydrolyze it. The S. coelicolor SC4828 protein has been crystallized with Ca2+ ions and phosphomethylphosphonic acid guanosyl ester. The SA1684 protein has amino acid sequence similarity with SC4828 protein and hydrolytic activity against NDPs in the presence of Mn2+ ions but not in the presence of Ca2+ ions. Further studies on the biochemical characteristics of SC4828 are needed to compare the functions of SA1684 and SC4828. A BLASTP search revealed that SA1684 or SC4828 are conserved among various pathogenic bacteria and non-pathogenic bacteria, including Bacillus, Staphylococcus, Listeria, Lactobacillus, Streptococcus, Mycoplasma, Frankia, Brachybacterium, Salinispora, Saccharopolyspora, Actinosynnema, and Streptomyces. These points might be important clues toward understanding the molecular functions of this novel nucleoside diphosphatase family and the physiological role beyond bacterial virulence. Eukaryotic nucleoside diphosphatases called apyrase, ecto-ATPase, and ATP-diphosphohydrolase have hydrolyzing activity against NTPs and NDPs in the presence of Ca2+ and Mg2+ ions (45). A few bacterial counterparts for apyrase have been discovered in Sulfolobus acidocaldarius (46), Shigella flexneri (47, 48), enteroinvasive E. coli (49), and Legionella pneumophila (50). The S. acidocaldarius enzyme hydrolyzes ATP and ADP in the absence of divalent cations (46). The L. pneumophila enzyme hydrolyzes ATP, GTP, ADP, and GDP in the presence of Mg2+, Ca2+, and Zn2+ ions. These biochemical characteristics of the apyrase family proteins differ from those of SA1684 and the primary sequences of apyrase and SA1684 also differ, indicating that the SA1684 protein could be a promising target of antimicrobial chemotherapy.
In this study, we identified five novel virulence genes, including the SA1684 gene, from among eight hypothetical genes using the silkworm infection model. Further investigation of these novel virulence factors will enhance our understanding of the bacterial virulence regulatory system.
Experimental Procedures
Bacterial Strains and Growth Conditions
The E. coli JM109 strain was used as the plasmid host. E. coli strains transformed with plasmids were aerobically cultured at 37 °C in Luria-Bertani broth containing 25 μg/ml chloramphenicol. S. aureus strains were aerobically cultured in tryptic soy broth at 37 °C, and 12.5 μg/ml chloramphenicol was added when culturing transformants with plasmids. The bacterial strains and plasmids used in this study are listed in Table 4.
TABLE 4.
List of bacterial strains and plasmids used
The following abbreviations are used: Erm, erythromycin; Tet, tetracycline; Cm, chloramphenicol; Km, kanamycin.
| Strain or plasmid | Genotypes or characteristics | Source or ref. |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | NCTC8325-4, restriction mutant | 34 |
| M0003 | RN4220 SA0003::ermAM; Ermr | This study |
| M0649 | RN4220 SA0649::ermAM; Ermr | This study |
| M0873 | RN4220 SA0873::ermAM; Ermr | This study |
| M0949 | RN4220 SA0949::ermAM; Ermr | This study |
| M1108 | RN4220 SA1108::ermAM; Ermr | This study |
| M1125 | RN4220 SA1125::ermAM; Ermr | This study |
| M1292 | RN4220 SA1292::ermAM; Ermr | This study |
| M1684 | RN4220 SA1684::ermAM; Ermr | This study |
| CK3 | RN4220 Δagr::tetM; Tetr | 16 |
| CK3D1684 | RN4220 Δagr::tetM, SA1684::ermAM; Tetr, Ermr | This study |
| E. coli | ||
| JM109 | Host strain for cloning | Takara Bio |
| BL21(DE3)-RIPL | Host strain for protein overproduction | Agilent Technologies |
| Plasmids | ||
| pKOR3a | Vector for allelic replacement in S. aureus, Cmr | 54 |
| pND50 | S. aureus-E. coli shuttle vector; Cmr | 32 |
| pSA1684 | pND50 with intact SA1684 | This study |
| pY88A | pND50 with SA1684 Y88A | This study |
| pD106A | pND50 with SA1684 D106A | This study |
| pD123A/E124A | pND50 with SA1684 D123A/E124A | This study |
| pET28b | E. coli vector for protein overproduction; Kmr | Novagen |
| pOp-SA1684 | pET28b with intact SA1684 | This study |
| pOp-Y88A | pET28b with SA1684 Y88A | This study |
| pOp-D106A | pET28b with SA1684 D106A | This study |
| pOp-D123A/E124A | pET28b with SA1684 D123A/E124A | This study |
| pCK5000 | pND50 with luc+ gene from pGL3 | 19 |
| pCK5001 | pCK5000 with agr P2 promoter from RN4220 | 19 |
| pCK5002 | pCK5000 with agr P3 promoter from RN4220 | 19 |
| pCK5003 | pCK5000 with hla promoter from RN4220 | 19 |
DNA Manipulation
Transformation of S. aureus with plasmids was performed by electroporation (51). Phage transduction was performed using phage 80α (52). Transformation of E. coli, extraction of plasmid DNA, PCR, and Southern blot hybridization were performed according to previously reported methods (53). Genomic DNA of S. aureus cells was extracted using a QIAamp DNA blood kit (Qiagen) after digesting the cell wall components using lysostaphin (Wako Chemicals, Tokyo, Japan).
Construction of S. aureus Gene-deletion Mutants by Double Crossover Homologous Recombination
A vector for allelic replacement in S. aureus, pKOR1 (24), was modified by removing the ccdB gene that is toxic to normal E. coli cells, resulting in pKOR3a (54). The DNA regions (∼800 bp) upstream and downstream of the target gene were amplified by PCR using primer pairs (Table 5) and genomic DNA of RN4220 strain as a template. The two amplified DNA fragments and a DNA fragment containing the erythromycin-resistant gene were spliced together by overlapped extension PCR. The amplified fragment was inserted into the SmaI site of pKOR3a to obtain the targeting vector. S. aureus RN4220 strain was transformed with the targeting vector and cultured at 43 °C, resulting in integration of the targeting vector into the chromosome. The strain was cultured on agar plates containing anhydrotetracycline to kill the bacteria harboring the plasmid, and the developing colony was examined for sensitivity against chloramphenicol and resistance against erythromycin. The deletion was transferred to the RN4220 strain by phage 80α, resulting in a new erythromycin-resistant mutant. The gene deletion was confirmed by Southern blot analysis.
TABLE 5.
Primers used in this study
F indicates forward; R indicates reverse.
| Primer | Sequence |
|---|---|
| SA0003L-F | ATGCACAGCGACTGACTCAC |
| SA0003L-R | ATTCTTGAAGACGAATGTCACAAATTTCATTTAAAATAGAGG |
| SA0003R-F | TTTGTAAATTTGGAATTAATCATTCATCAAGGTGAACA |
| SA0003R-R | GGACAAAGCCGTTGTACGTT |
| SA0649L-F | TTGGAAAAGTGCCCCAAATA |
| SA0649L-R | ATTCTTGAAGACGAATGATCAATCCCCTTTATTTTAATATGT |
| SA0649R-F | TTTGTAAATTTGGAATCAATAGCACTTTACTTTTTAGTTGAA |
| SA0649R-R | CCTAGTCCAAATGGCAGCAT |
| SA0873L-F | TGATGCGATCATTGTTGGAT |
| SA0873L-R | ATTCTTGAAGACGAATTCCTCCTTTGTTTAACCTATTGC |
| SA0873R-F | TTTGTAAATTTGGAAAGATAAATGGAAAGTTATTGAAACG |
| SA0873R-R | GTGGCAAACAAATGGGCTAT |
| SA0949L-F | GGTTATTCAGAAGCGCAAGC |
| SA0949L-R | ATTCTTGAAGACGAACCTATGTTCATTTATTTTTCCACCT |
| SA0949R-F | TTTGTAAATTTGGAAGCGACAGCTTCATATTTATAGGG |
| SA0949R-R | TAACCATGCGTCCTTCAACA |
| SA1108L-F | CATGCAGCAGCATACGTTTT |
| SA1108L-R | ATTCTTGAAGACGAAACAGGCCTCCCTTTTTGG |
| SA1108R-F | TTTGTAAATTTGGAAAACGTGATGAGGAGGAAAAA |
| SA1108R-R | AACACATGCACCAACAGCAT |
| SA1125L-F | GGTGAGGTTCACCATGACAG |
| SA1125L-R | ATTCTTGAAGACGAACGTTTTCAATTCATAGCCTCCT |
| SA1125R-F | TTTGTAAATTTGGAACAGAAATAAATTAGTGAGAAATGAGGA |
| SA1125R-R | CCGTTGGTATTAGCGATTTGA |
| SA1292L-F | TCCTTTATGGGCACAACACA |
| SA1292L-R | ATTCTTGAAGACGAATTCTACGTTCTCCTTATATTGCATTTC |
| SA1292R-F | TTTGTAAATTTGGAAAAAATCGATTCGAAAGAAAGTGA |
| SA1292R-R | GACGCATTGGCACTAATTCA |
| SA1684L-F | ACAATCGACGAGGAAATCGT |
| SA1684L-R | ATTCTTGAAGACGAAACTCCACACCACCTTCTGTT |
| SA1684R-F | TTTGTAAATTTGGAATTTTATCAAAGTTTGGAAAGAACG |
| SA1684R-R | CGCTTCATTGTCTTCAATCG |
Silkworm Infection Experiment
Silkworms were raised in our laboratory (12). Fifth instar larvae were fed an antibiotic-free diet (Sysmex Corp., Kobe, Japan) for 1 day and then injected with 50 μl of bacterial solution using a 1-ml syringe equipped with a 27-gauge needle. Surviving silkworms were counted at 24 h post-infection. Infection experiments were performed at 27 °C.
Colony Spreading Assay
Tryptic soy broth containing 0.24% agar (Nacalai Tesque, Kyoto, Japan) was autoclaved, and the sterile medium was poured into 8-cm plates (30). The plates were dried for 20 min in a safety cabinet and then spotted with 2 μl of S. aureus overnight culture and dried further for 20 min. The plates were incubated at 37 °C for 8 h.
Measurement of Hemolysin and Nuclease Production
To measure S. aureus hemolysin production, S. aureus overnight cultures were spotted onto tryptic soy broth agar plates containing 5% sheep erythrocytes and incubated overnight. Quantitative measurement of the hemolytic activity was performed according to a previous method (19). Briefly, 2-fold serial diluted supernatants of S. aureus overnight cultures were mixed with sheep blood erythrocytes for 1 h at 37 °C in a 96-well microplate. The plate was centrifuged, and absorbance of the supernatants at 450 nm was measured. The reciprocal of the dilution that caused 50% lysis were determined as the hemolytic unit.
To measure S. aureus nuclease production, S. aureus overnight cultures were spotted onto a DNA agar plate (Eiken, Japan) and incubated overnight. The plate was reacted with 1.5 n HCl, and the clear zone around the bacterial colony was measured.
Reporter Assay
S. aureus RN4220 strain and the gene-deletion mutants were transformed with reporter plasmids (Table 4). They were cultured to A600 = 1, collected by centrifugation, and lysed in a buffer (20 mm potassium phosphate buffer (pH 7.8), 0.04% Triton X-100, 100 μm DTT, 10 μg/ml lysostaphin, 1 tablet of protease inhibitor/50 ml (Complete, Roche Applied Science). Cell lysate supernatant (100 μl) was incubated with an equal volume of luciferase substrate (Promega, Madison, WI), and luminescence was measured using a luminometer (Berthold Technologies, Bad Wildbad, Germany). The promoter activity was calculated as luminescence units/mg of protein.
Analysis of Extracellular Proteins, Cell Wall Proteins, and Cytosol/Membrane Proteins
S. aureus extracellular proteins, cell wall proteins, and cytosol/membrane proteins were fractionated according to a previous method (19, 55) with minor modifications. S. aureus overnight culture (50 μl) was inoculated into 5 ml of fresh tryptic soy broth and aerobically cultured for 15 h at 37 °C. The culture was centrifuged at 20,400 × g for 5 min, and the supernatant was mixed with TCA (final concentration, 10%). The sample was incubated on ice for 30 min and centrifuged at 20,400 × g for 25 min at 4 °C. The precipitate was washed twice with ice-cold ethanol. The precipitate was dissolved in SDS sample buffer and analyzed by SDS-PAGE. To obtain the cell wall proteins and cytosol/membrane proteins, the 15-h culture was centrifuged at 20,400 × g for 5 min, and the precipitated bacterial cells were suspended in a digestion solution (50 mm Tris-HCl (pH 7.5), 145 mm NaCl, 30% raffinose, lysostaphin 200 μg/ml, DNase I 10 μg/ml) and incubated at 37 °C for 30 min. The sample was centrifuged at 9560 × g for 10 min at 4 °C. The precipitate was dissolved in SDS sample buffer and analyzed as cytosol/membrane proteins. The centrifuged supernatant was mixed with TCA (final concentration, 10%) and centrifuged at 20,400 × g for 25 min at 4 °C. The precipitate was washed with ice-cold ethanol, dissolved in SDS sample buffer, and analyzed as cell wall proteins.
Purification of SA1684 Protein
E. coli BL21(DE3)-RIPL strain was transformed with pET28b or pOp-SA1684 and cultured in Terrific broth containing 1 m sorbitol and 10 mm betaine at 37 °C overnight. Ten milliliters of the overnight culture was inoculated into 1 liter of the same fresh medium and cultured to A600 = 0.4 at 37 °C. The culture was supplemented with isopropyl β-d-1-thiogalactopyranoside (final concentration, 0.5 mm) and cultured overnight at 16 °C. The culture was centrifuged at 12,020 × g for 10 min, and the precipitated bacterial cells were suspended in a lysis buffer (20 mm Tris-HCl (pH 7.2), 500 mm NaCl, 5 mm imidazole) and frozen in liquid nitrogen. The frozen cells were dissolved, lysed by sonication, and centrifuged at 91,287 × g for 1 h. The supernatant was mixed with nickel affinity resin (ProBond resin, Life Technologies, Inc.) for 1 h at 4 °C. The resin was washed with a wash buffer (20 mm Tris-HCl (pH 7.2), 500 mm NaCl, 67.2 mm imidazole). Protein was eluted from the resin with an elution buffer (20 mm Tris-HCl (pH 7.2), 500 mm NaCl, 1 m imidazole). The protein concentration was determined by the Bradford assay.
Measurement of Nucleoside Diphosphate Hydrolysis
Nucleoside diphosphate was mixed with protein solution in a reaction buffer (20 mm HEPES-KOH (pH 7.5), 50 mm NaCl, 0.5 mm MnCl2). The reaction product was analyzed by TLC on PEI-Cellulose F plates (Merck) by a solvent (saturated ammonium sulfate:3 m sodium acetate:isopropyl alcohol = 40:3:1). The reaction product was visualized under UV light (254 nm). To quantitatively measure the NDP hydrolysis, the reaction was stopped by EDTA (final concentration: 5 mm), and the free phosphate in the reaction sample was measured by the malachite green-based method (56). KH2PO4 was used as a standard. The kinetic parameters of the enzyme reaction were determined by nonlinear regression analysis using Graph Pad Prism version 5.0c.
Quantitative Reverse Transcription-PCR Analysis
S. aureus overnight cultures (50 μl) were inoculated into 5 ml of tryptic soy broth and cultured to A600 = 1 at 37 °C. Total RNA was extracted using RNAprotect bacteria reagent (Qiagen) and an RNeasy mini kit (Qiagen). cDNA was synthesized from total RNA using a random hexamer and MultiScribe Reverse Transcriptase (Applied Biosystems). Quantitative PCR analysis was performed as described previously (57).
RNA Sequence Analysis
Total RNA was extracted as described above and RNA-seq template was prepared using an Agilent strand-Specific RNA prep kit (Agilent) without poly(A) selection. Sequencing was performed using a Hiseq2500 platform (Illumina). At least 10 million sequences of 36-base single end reads were generated per sample. The data were analyzed using CLC Genomics Workbench software, version 8.5.1 (CLC Bio, Aarhus, Denmark). The number of reads per kilobase per million mapped reads was compared between the parent strain and the SA1684-deletion mutant or between the SA1684-deletion mutant and the complemented strain. Experiments were performed three times independently, and the differential gene expression was statistically analyzed using “Empirical Analysis of DGE (differential gene expression).” Genes with more than 2-fold differential expression and with a false discovery rate p value less than 0.05 were identified.
Gene enrichment analysis was performed according to the previously described method (58, 59). Normalized RNA sequence data of the parent strain and the SA1684-deletion mutant were formatted as a .gct and .chip file. The S. aureus NCTC8325 genome contains 2844 genes, of which 935 genes are categorized in 106 pathways in the KEGG database and were formatted as a .gmt file. The analysis was performed using Gene Set Enrichment Analysis (GESA) software. Differences in the pathways were considered significant if the normalized p value was less than 0.05.
Author Contributions
C. K. conceived and coordinated the study and wrote the paper. Y. Saito, H. M., and C. K. designed, performed, and analyzed the experiments in Figs. 1 and 2. K. I., H. K., H. R., A. M., K. S, and C. K. designed, performed, and analyzed the experiments in Figs. 3–6 and Table 2. H. R., Y. Suzuki, and C. K. performed and analyzed the experiments in Tables 5 and supplemental Tables S1 and S2. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by Grants-in-aid for Scientific Research 15H04727 and 15H05783, Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant 221S0002, and in part by The KANAE Foundation and the Genome Pharmaceutical Institute. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Tables S1 and S2.
- MRSA
- methicillin-resistant S. aureus
- (p)ppGpp
- guanosine pentaphosphate
- GDPβS
- guanosine 5′-O-2-(thio)diphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
References
- 1. Novick R. P. (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 [DOI] [PubMed] [Google Scholar]
- 2. Fournier B., Klier A., and Rapoport G. (2001) The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41, 247–261 [DOI] [PubMed] [Google Scholar]
- 3. Giraudo A. T., Cheung A. L., and Nagel R. (1997) The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168, 53–58 [DOI] [PubMed] [Google Scholar]
- 4. Besier S., Ludwig A., Ohlsen K., Brade V., and Wichelhaus T. A. (2007) Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 297, 217–225 [DOI] [PubMed] [Google Scholar]
- 5. Chatterjee I., Kriegeskorte A., Fischer A., Deiwick S., Theimann N., Proctor R. A., Peters G., Herrmann M., and Kahl B. C. (2008) In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 190, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kriegeskorte A., Block D., Drescher M., Windmüller N., Mellmann A., Baum C., Neumann C., Lorè N. I., Bragonzi A., Liebau E., Hertel P., Seggewiss J., Becker K., Proctor R. A., Peters G., and Kahl B. C. (2014) Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio 5, e01447–01414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stenz L., Francois P., Whiteson K., Wolz C., Linder P., and Schrenzel J. (2011) The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 62, 123–139 [DOI] [PubMed] [Google Scholar]
- 8. Srivatsan A., and Wang J. D. (2008) Control of bacterial transcription, translation and replication by (p) ppGpp. Curr. Opin. Microbiol. 11, 100–105 [DOI] [PubMed] [Google Scholar]
- 9. Geiger T., and Wolz C. (2014) Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 304, 150–155 [DOI] [PubMed] [Google Scholar]
- 10. Karaolis D. K., Rashid M. H., Chythanya R., Luo W., Hyodo M., and Hayakawa Y. (2005) c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell-cell interactions and biofilm formation. Antimicrob. Agents Chemother. 49, 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ryan R. P. (2013) Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 159, 1286–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaito C., Akimitsu N., Watanabe H., and Sekimizu K. (2002) Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb. Pathog. 32, 183–190 [DOI] [PubMed] [Google Scholar]
- 13. Kaito C., and Sekimizu K. (2007) A silkworm model of pathogenic bacterial infection. Drug Discov. Ther. 1, 89–93 [PubMed] [Google Scholar]
- 14. Kaito C., Yoshikai H., and Sekimizu K. (2012) Utilization of a silkworm model for understanding host-pathogen interactions. Isj-Invert. Surviv. J. 9, 163–168 [Google Scholar]
- 15. Miyazaki S., Matsumoto Y., Sekimizu K., and Kaito C. (2012) Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol. Lett. 326, 116–124 [DOI] [PubMed] [Google Scholar]
- 16. Kaito C., Kurokawa K., Matsumoto Y., Terao Y., Kawabata S., Hamada S., and Sekimizu K. (2005) Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56, 934–944 [DOI] [PubMed] [Google Scholar]
- 17. Kaito C., Morishita D., Matsumoto Y., Kurokawa K., and Sekimizu K. (2006) Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol. Microbiol. 62, 1601–1617 [DOI] [PubMed] [Google Scholar]
- 18. Nagata M., Kaito C., and Sekimizu K. (2008) Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem. 283, 2176–2184 [DOI] [PubMed] [Google Scholar]
- 19. Matsumoto Y., Kaito C., Morishita D., Kurokawa K., and Sekimizu K. (2007) Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene. Infect. Immun. 75, 1964–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsumoto Y., Xu Q., Miyazaki S., Kaito C., Farr C. L., Axelrod H. L., Chiu H. J., Klock H. E., Knuth M. W., Miller M. D., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Sekimizu K., and Wilson I. A. (2010) Structure of a virulence regulatory factor CvfB reveals a novel winged helix RNA binding module. Structure 18, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikuo M., Kaito C., and Sekimizu K. (2010) The cvfC operon of Staphylococcus aureus contributes to virulence via expression of the thyA gene. Microb. Pathog. 49, 1–7 [DOI] [PubMed] [Google Scholar]
- 22. Kyuma T., Kimura S., Hanada Y., Suzuki T., Sekimizu K., and Kaito C. (2015) Ribosomal RNA methyltransferases contribute to Staphylococcus aureus virulence. FEBS J. 282, 2570–2584 [DOI] [PubMed] [Google Scholar]
- 23. Kyuma T., Kizaki H., Ryuno H., Sekimizu K., and Kaito C. (2015) 16S rRNA methyltransferase KsgA contributes to oxidative stress resistance and virulence in Staphylococcus aureus. Biochimie 119, 166–174 [DOI] [PubMed] [Google Scholar]
- 24. Bae T., and Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 [DOI] [PubMed] [Google Scholar]
- 25. Li D., Liu C., Liang Y. H., Li L. F., and Su X. D. (2008) Crystal structure of B. subtilis YjcG characterizing the YjcG-like group of 2H phosphoesterase superfamily. Proteins 72, 1071–1076 [DOI] [PubMed] [Google Scholar]
- 26. Nasr F., and Filipowicz W. (2000) Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. Nucleic Acids Res. 28, 1676–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., and Inouye M. (2002) MazG, a nucleoside triphosphate pyrophosphohydrolase, interacts with Era, an essential GTPase in Escherichia coli. J. Bacteriol. 184, 5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gross M., Marianovsky I., and Glaser G. (2006) MazG–a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59, 590–601 [DOI] [PubMed] [Google Scholar]
- 29. Lee S., Kim M. H., Kang B. S., Kim J. S., Kim G. H., Kim Y. G., and Kim K. J. (2008) Crystal structure of Escherichia coli. MazG, the regulator of nutritional stress response. J. Biol. Chem. 283, 15232–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaito C., and Sekimizu K. (2007) Colony spreading in Staphylococcus aureus. J. Bacteriol. 189, 2553–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., and Moghazeh S. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaito C., Kurokawa K., Hossain M. S., Akimitsu N., and Sekimizu K. (2002) Isolation and characterization of temperature-sensitive mutants of the Staphylococcus aureus dnaC gene. FEMS Microbiol. Lett. 210, 157–164 [DOI] [PubMed] [Google Scholar]
- 33. Manna A. C., and Cheung A. L. (2006) Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188, 4288–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., and Schlievert P. (1988) Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ueda T., Kaito C., Omae Y., and Sekimizu K. (2011) Sugar-responsive gene expression and the agr system are required for colony spreading in Staphylococcus aureus. Microb. Pathog. 51, 178–185 [DOI] [PubMed] [Google Scholar]
- 36. Khemici V., Prados J., Linder P., and Redder P. (2015) Decay-initiating endoribonucleolytic cleavage by RNase Y is kept under tight control via sequence preference and sub-cellular localisation. PLoS Genet. 11, e1005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Numata S., Nagata M., Mao H., Sekimizu K., and Kaito C. (2014) CvfA protein and polynucleotide phosphorylase act in an opposing manner to regulate Staphylococcus aureus virulence. J. Biol. Chem. 289, 8420–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kohler C., von Eiff C., Liebeke M., McNamara P. J., Lalk M., Proctor R. A., Hecker M., and Engelmann S. (2008) A defect in menadione biosynthesis induces global changes in gene expression in Staphylococcus aureus. J. Bacteriol. 190, 6351–6364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., Brown E. L., Zagursky R. J., Shlaes D., and Projan S. J. (2001) Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183, 7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheung G. Y., Wang R., Khan B. A., Sturdevant D. E., and Otto M. (2011) Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagarajan V., and Elasri M. O. (2007) SAMMD: Staphylococcus aureus microarray meta-database. BMC Genomics 8, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Majerczyk C. D., Sadykov M. R., Luong T. T., Lee C., Somerville G. A., and Sonenshein A. L. (2008) Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., Bodi K., and Sonenshein A. L. (2010) Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roux A., Todd D. A., Velázquez J. V., Cech N. B., and Sonenshein A. L. (2014) CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol. 196, 1184–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yegutkin G. G. (2008) Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim. Biophys. Acta 1783, 673–694 [DOI] [PubMed] [Google Scholar]
- 46. Amano T., Wakagi T., and Oshima T. (1993) An ecto-enzyme from Sulfolobus acidocaldarius strain 7 which catalyzes hydrolysis of inorganic pyrophosphate, ATP, and ADP: purification and characterization. J. Biochem. 114, 329–333 [DOI] [PubMed] [Google Scholar]
- 47. Bhargava T., Datta S., Ramachandran V., Ramakrishnan R., Roy R. K., Sankaran K., and Subrahmanyam Y. V. (1995) Virulent Shigella codes for a soluble apyrase–identification, characterization and cloning of time gene. Curr. Sci. 68, 293–300 [Google Scholar]
- 48. Sarli S., Nicoletti M., Schippa S., Del Chierico F., Santapaola D., Valenti P., and Berlutti F. (2005) Ala160 and His116 residues are involved in activity and specificity of apyrase, an ATP-hydrolysing enzyme produced by enteroinvasive Escherichia coli. Microbiology 151, 2853–2860 [DOI] [PubMed] [Google Scholar]
- 49. Berlutti F., Casalino M., Zagaglia C., Fradiani P. A., Visca P., and Nicoletti M. (1998) Expression of the virulence plasmid-carried apyrase gene (apy) of enteroinvasive Escherichia coli and Shigella flexneri is under the control of H-NS and the VirF and VirB regulatory cascade. Infect. Immun. 66, 4957–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sansom F. M., Newton H. J., Crikis S., Cianciotto N. P., Cowan P. J., d'Apice A. J., and Hartland E. L. (2007) A bacterial ecto-triphosphate diphosphohydrolase similar to human CD39 is essential for intracellular multiplication of Legionella pneumophila. Cell Microbiol. 9, 1922–1935 [DOI] [PubMed] [Google Scholar]
- 51. Schenk S., and Laddaga R. A. (1992) Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73, 133–138 [DOI] [PubMed] [Google Scholar]
- 52. Novick R. P. (1991) Genetic systems in staphylococci. Methods Enzymol. 204, 587–636 [DOI] [PubMed] [Google Scholar]
- 53. Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Kaito C., Hirano T., Omae Y., and Sekimizu K. (2011) Digestion of extracellular DNA is required for giant colony formation of Staphylococcus aureus. Microb. Pathog. 51, 142–148 [DOI] [PubMed] [Google Scholar]
- 55. Hartford O., Francois P., Vaudaux P., and Foster T. J. (1997) The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 56. Carter S. G., and Karl D. W. (1982) Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7, 7–13 [DOI] [PubMed] [Google Scholar]
- 57. Kaito C., Saito Y., Nagano G., Ikuo M., Omae Y., Hanada Y., Han X., Kuwahara-Arai K., Hishinuma T., Baba T., Ito T., Hiramatsu K., and Sekimizu K. (2011) Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 7, e1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hansen J. J., Huang Y., Peterson D. A., Goeser L., Fan T. J., Chang E. B., and Sartor R. B. (2012) The colitis-associated transcriptional profile of commensal Bacteroides thetaiotaomicron enhances adaptive immune responses to a bacterial antigen. PLoS ONE 7, e42645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.