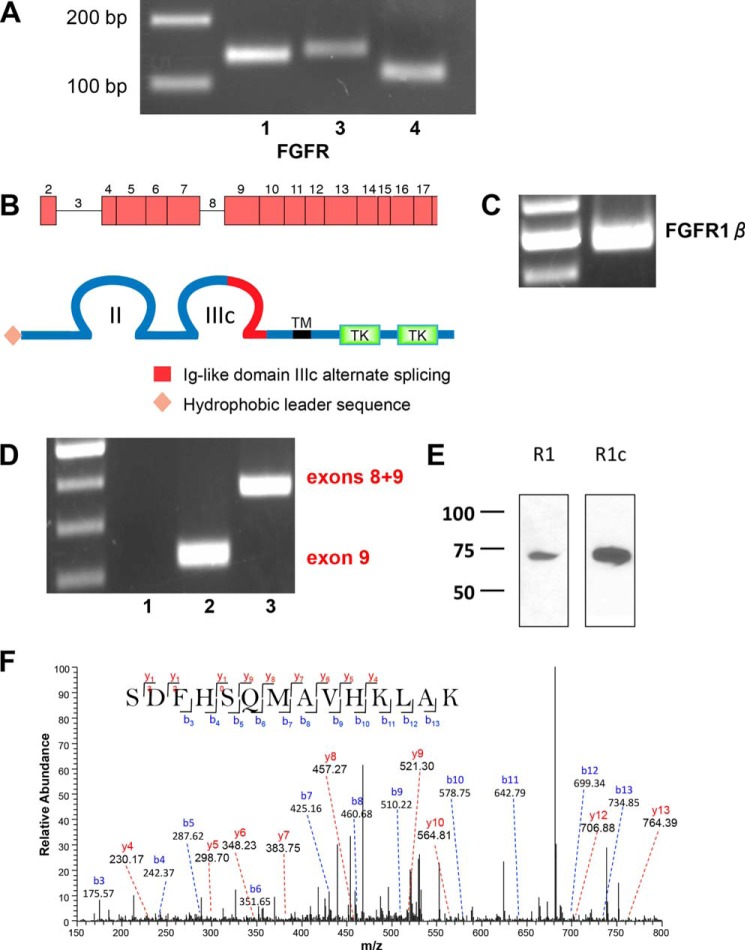
FIGURE 2.
FGFR mRNA and protein expression in RPTECs. A, representative 3% agarose gel of FGFR expression in RPTECs determined by RT-PCR (Table 2). 200- and 100-bp markers are shown in the left lane. B, schematic representations of the FGFR1 gene and protein structure. The FGFR-β1c splice variant harbors two extracellular immunoglobulin-like domains (II and III) with alternatively spliced exon 9 in loop III, a transmembrane domain (TM), and two intracellular tyrosine kinase domains (TK). C, FGFR1-β splice variant in a characteristic 2% agarose gel with ladder in the left lane. The 500-bp PCR product is consistent with the β-splice variant lacking exon 3. D, FGFR1b/c splice variants. Lane 1, FGFR1b; lane 2, FGFR1c; lane 3, mRNA containing both exons 8 and 9. Lane 3 PCR was performed using the forward primer for FGFR1b and the reverse primer for FGFR1c (Table 1). An illustrative 2% agarose gel is shown with a 100-bp ladder in the left lane. The PCR product in lane 2 is consistent with the exon 9 FGFR1c splice variant. Lane 3 shows an mRNA species containing both exons 8 and 9. E, FGFR1c is the primary species of FGFR1 detected by immunoblotting. FGFR1 and FGFR1c in RPTECs were detected by non-selective and isotype-selective antisera, respectively, as described under “Experimental Procedures.” The results are illustrative of n = 3 independent experiments. F, MS/MS spectrum for the identified specific peptide 355SDFHSQMAVHKLAK368 from FGFR1c. The peak heights show the relative abundances of the corresponding fragmentation ions, with the annotation of the identified matched N terminus-containing b ions in blue and the C terminus-containing y ions in red. Charge state: +3, observed m/z: 538.93969, theoretical m/z: 538.94008, precursor mass error: −0.72, Xcorr: 0.719.