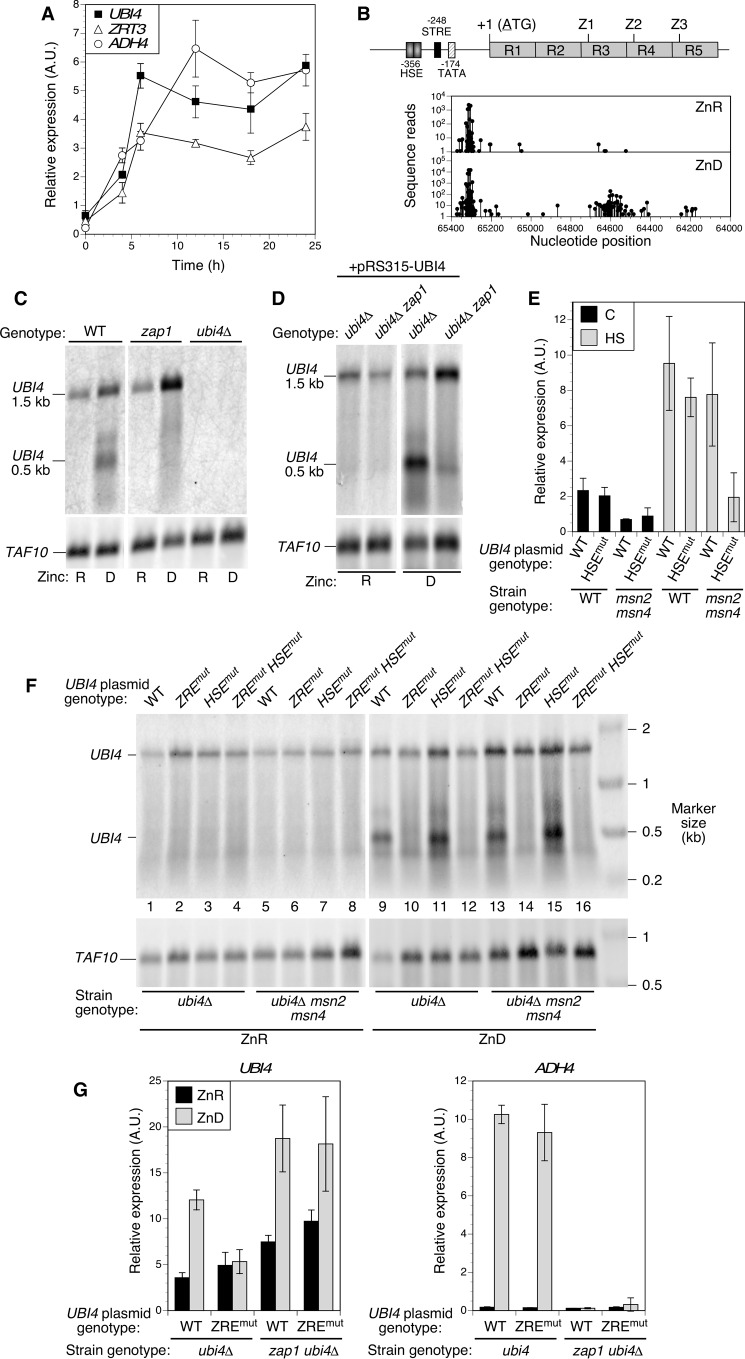
FIGURE 6.
Zap1 induces UBI4 expression in zinc-deficient cells via an intragenic promoter. A, UBI4 induction parallels Zap1 target gene expression in zinc-deficient conditions. Log phase cultures of wild-type and ubi4Δ mutant cells grown in zinc-replete medium (LZM + 100 μm ZnCl2) were used to inoculate zinc-deficient medium at time 0 (LZM + 1 μm ZnCl2). Cells were harvested at the indicated times, and expression of the indicated genes was assayed by RT-qPCR. Each data point is the mean of three replicates, and the error bars denote ±1 S.D. B, location of potential Zap1 binding sites (ZREs) and transcription start sites in UBI4. The structure of the UBI4 gene is shown with its previously mapped promoter elements for Hsf1 (HSE)- and Msn2/Msn4 (STRE)-dependent regulation and TATA box. R1–R5 represent the ubiquitin repeats, and the locations of the potential intragenic ZREs are marked (Z1–Z3). The graphs below show the transcription start sites mapped by RLM-RACE using mRNA from zinc-replete (ZnR; LZM + 100 μm ZnCl2) and zinc-deficient (ZnD; LZM + 1 μm ZnCl2) cells. C and D, short 0.5-kb UBI4 transcripts are dependent on zinc supply and Zap1. Northern blotting analysis of UBI4 expressed from the chromosomal locus (C) or a UBI4 plasmid bearing the full promoter (pRS315-UBI4; D) is shown. For C, wild-type, zap1Δ, and ubi4Δ haploid strains were transformed with pGK-ZRT1 to allow growth of zap1Δ mutants in low zinc and then grown in zinc-replete (R; LZM + 100 μm ZnCl2) or zinc-deficient (D; LZM + 1 μm ZnCl2) conditions prior to RNA extraction. For D, zap1Δ (CWM260) and zap1Δ ubi4Δ (CWM276) strains were transformed with both pRS315-UBI4 and pGK-ZRT1 prior to growth in zinc-deficient or -replete medium and RNA extraction. For C and D, UBI4 mRNA was detected using a probe specific for the 3′-UTR, and TAF10 was also detected as a loading control. E, mutation of the UBI4 HSE blocks UBI4 induction by heat shock but only in an msn2Δ msn4Δ mutant background. Wild-type (BY4741) or msn2Δ msn4Δ (DBY9435) strains were transformed with pRS315-UBI4 (wild-type UBI4) or pRS315-UBI4HSEmut (HSE mutant). Cells were grown to log phase at 25 °C in SD medium or subjected to heat shock (HS; 20 min at 39 °C) before harvesting. Total RNA was isolated and assayed for UBI4 expression using RT-qPCR. Each data point is the mean of three replicates, and the error bars denote ±1 S.D. F, the UBI4 ZRE is essential for production of intragenic transcripts. Yeast strains ubi4Δ (CWM260) and ubi4Δ msn2Δ msn4Δ (CWM274) were transformed with either the wild-type UBI4 plasmid (pRS315-UBI4), a plasmid in which the three intragenic ZREs were mutated (pRS315-UBI4ZREmut), the HSE mutant plasmid (pRS315-UBI4HSEmut), or a plasmid in which both the ZREs and the HSE were mutated (pRS315-UBI4HSEmutZREmut). Cells were grown and analyzed by Northern blotting as described for C and D. G, the ZREs are required for UBI4 induction in zinc deficiency. Mutant ubi4Δ (CWM260) and ubi4Δ zap1Δ (CWM276) cells were transformed with pGK-ZRT1 together with either the wild-type UBI4 plasmid (pRS315-UBI4) or the ZRE mutant plasmid (pRS315-UBI4ZREmut). Cells were grown to log phase in zinc-replete (ZnR; LZM + 100 μm ZnCl2) or zinc-deficient (ZnD; LZM + 1 μm ZnCl2) medium prior to RNA extraction. Quantitative RT-PCR was used to measure total UBI4 transcripts using primers specific to the fifth ubiquitin repeat that is present in both full-length and short transcripts. A Zap1 target gene (ADH4) was also detected as a positive control for loss of Zap1 function. Each data point is the mean of three replicates, and the error bars denote ±1 S.D. A.U., arbitrary units.