Abstract
Spider dragline silk is a natural polymer harboring unique physical and biochemical properties that make it an ideal biomaterial. Artificial silk production requires an understanding of the in vivo mechanisms spiders use to convert soluble proteins, called spidroins, into insoluble fibers. Controlled dimerization of the spidroin N-terminal domain (NTD) is crucial to this process. Here, we report the crystal structure of the Nephila clavipes major ampullate spidroin NTD dimer. Comparison of our N. clavipes NTD structure with previously determined Euprosthenops australis NTD structures reveals subtle conformational alterations that lead to differences in how the subunits are arranged at the dimer interface. We observe a subset of contacts that are specific to each ortholog, as well as a substantial increase in asymmetry in the interactions observed at the N. clavipes NTD dimer interface. These asymmetric interactions include novel intermolecular salt bridges that provide new insights into the mechanism of NTD dimerization. We also observe a unique intramolecular “handshake” interaction between two conserved acidic residues that our data suggest adds an additional layer of complexity to the pH-sensitive relay mechanism for NTD dimerization. The results of a panel of tryptophan fluorescence dimerization assays probing the importance of these interactions support our structural observations. Based on our findings, we propose that conformational selectivity and plasticity at the NTD dimer interface play a role in the pH-dependent transition of the NTD from monomer to stably associated dimer as the spidroin progresses through the silk extrusion duct.
Keywords: crystal structure, dimerization, protein self-assembly, protein structure, structural biology, structure-function, x-ray crystallography, MaSp1A, spidroin, spider silk, structural plasticity, Nephila clavipes
Introduction
Spider silk is a naturally occurring polymer that has high elasticity, high tensile strength, and a biodegradable nature (1), which make it an ideal biomaterial for applications such as skin graft scaffolds (2), neuron regeneration (3), and cartilage repair (4). Dragline silk, one of the strongest types of silk, is produced in the major ampullate gland and is used as the web frame and lifeline for the spider (5). The proteins that form the core of dragline silk are termed major ampullate spidroins (MaSp),2 and they have three main domains: a long, flexible, highly repetitive central domain known as the repeat region flanked by smaller, globular, non-repetitive N-terminal and C-terminal domains (NTD and CTD, respectively) (6–8). As silk is being spun, the spidroins progress down a narrowing duct and experience a progressive decrease in pH and salt concentration that promotes a monomer to dimer transition of the NTD (9). This controlled oligomerization process serves to prevent precocious aggregation in the ampullate gland and also to promote fiber formation in the spinning duct (7).
Studies aimed at elucidating the structural basis for pH- and salt-dependent NTD dimerization have resulted in snapshots of the Euprosthenops australis NTD in both a monomeric (10) and dimeric (11) state, using NMR spectroscopy and x-ray crystallography, respectively. Based on these studies, a model for the monomer to dimer transition of the E. australis NTD (henceforth EaNTD) that depends on a pH-sensitive relay has been proposed. As the spidroins progress through the spinning duct, the decrease in salt concentration promotes intermolecular electrostatic interactions that align the monomer subunits into a weakly associated dimer. In particular, long-range electrostatic interactions between Lys65 and Asp40 have been shown to be crucial to the dimerization process. A concomitant decrease in pH to ∼6.5 results in protonation of two acidic residues (Glu79 and Glu119) (12), which promotes structural conversions within the NTD subunits that are also required to form this weakly associated dimer. As the pH decreases to ∼5.7 near the end of the spinning duct, protonation of a third acidic residue (Glu84) facilitates formation of the fully stable dimer (13). Notably, all of the key interactions at the dimer interface exhibit a high degree of symmetry, meaning that equivalent residues of each subunit engage in the same sets of contacts (11).
A more complete understanding of the molecular mechanisms governing pH-dependent dimerization of the NTD during silk fiber formation in vivo could facilitate artificial production of spider silk for use as a biomaterial. Further, silks produced from different spider species vary significantly in physical properties such as flexibility and tensile strength (6). Although currently unknown, the molecular basis underlying these differing properties could prove useful for generation of silks optimized for different applications. To gain insights into these processes, we determined the crystal structure of the Nephila clavipes NTD (henceforth NcNTD) to 2.02 Å resolution. Surprisingly, despite having high sequence identity, our structural analysis revealed a distinct asymmetry at the NcNTD dimer interface relative to its E. australis ortholog. Among these asymmetric interactions are novel intermolecular salt bridges at opposite ends of the dimer (Asp39-Lys65 and Asp40-Lys65) that provide new insights into the crucial role these residues play in NTD dimer formation. Our structure also reveals a unique intramolecular “handshake” interaction between Asp17 and Asp53, which contributes to the asymmetry seen in our structure and potentially adds an additional layer of complexity to the pH-sensitive relay mechanism for NTD dimerization proposed by Knight and colleagues (11, 13). We investigated the role these key salt bridge and handshake interactions play in dimer formation using a tryptophan fluorescence assay, and the results support our structural observations. Based on our findings, we propose that plasticity at the NTD dimer interface plays a role in the pH-dependent transition of the NTD from a loosely to stably associated dimer as the spidroin progresses through the spider's silk extrusion duct.
Results and Discussion
Overall Structure of the N. clavipes Major Ampullate Spidroin NTD
The sequence identity and overall domain organization of spidroins are highly conserved across spider species, as is the property of pH-dependent dimerization of the NTD as the protein progresses through the spinning duct (1, 7). Despite these many similarities, the major ampullate silks from various spider species display different physical qualities. For example, E. australis dragline silk displays high tensile strength, whereas Nephila edulis dragline silk is much more flexible (6). To further elucidate the biochemical mechanisms that mediate NTD dimerization and their potential impact on silk properties, we determined the crystal structure of the N. clavipes MaSp1A NTD to 2.02 Å resolution (Table 1) using the structure of the E. australis MaSp1 NTD (Protein Data Bank (PDB): 3LR2) (11) as the search model for molecular replacement. The model was refined to working and free R-values of 16.4 and 20.5%, respectively (Table 1).
TABLE 1.
Crystallographic data and refinement statistics
Parentheses indicate statistics for the high-resolution data bin for x-ray data.
| N. clavipes MaSp1A NTD | |
|---|---|
| Crystallographic data | |
| Source | APS 22 ID |
| Wavelength (Å) | 1.00 |
| Resolution limits (Å) | 58.5–2.02 (2.07–2.02) |
| Space group | P3121 |
| Unit cell (Å) a, b, c | 67.5, 67.5, 90.4 |
| Unit cell (°) α, β, γ | 90, 90, 120 |
| No. of observations | 17,4181 (12,661) |
| No. of reflections | 16,120 (1145) |
| Completeness (%) | 98.23 (94.32) |
| Mean I/σI | 21.73 (5.84) |
| Rmerge on Ia | 0.081 (0.60) |
| CC½b | 99.9 (93.5) |
| Refinement statistics | |
| Resolution Limits (Å) | 35.74–2.02 (2.09–2.02) |
| No. of reflections (work/free) | 15,808/1561 |
| Completeness (%) | 98.22 (94.6) |
| Cutoff Criteria I/σI | 0 |
| Protein/water atoms | 1844/120 |
| Rcrystc | 0.164 (0.169) |
| Rfree (10% of data) | 0.205 (0.233) |
| Bonds (Å)/Angles (°) | 0.004/0.724 |
| B-factors: protein/water (Å2) | 36.7/40.5 |
| Ramachandran plot statistics (%) | |
| Preferred regions | 100 |
| Outlier regions | 0 |
| MolProbity score | 0.77–100th percentile (n = 12,151, 2.02 Å ± 0.25Å) |
| PDB ID | 5IZ2 |
a Rmerge = Σhkl Σi|I(hkl)i − 〈I(hkl)〉|/ΣhklΣi 〈I(hkl)i〉.
b CC½, correlation coefficient.
c Rcryst = Σhkl |Fo(hkl) − Fc(hkl)|/Σhkl |Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
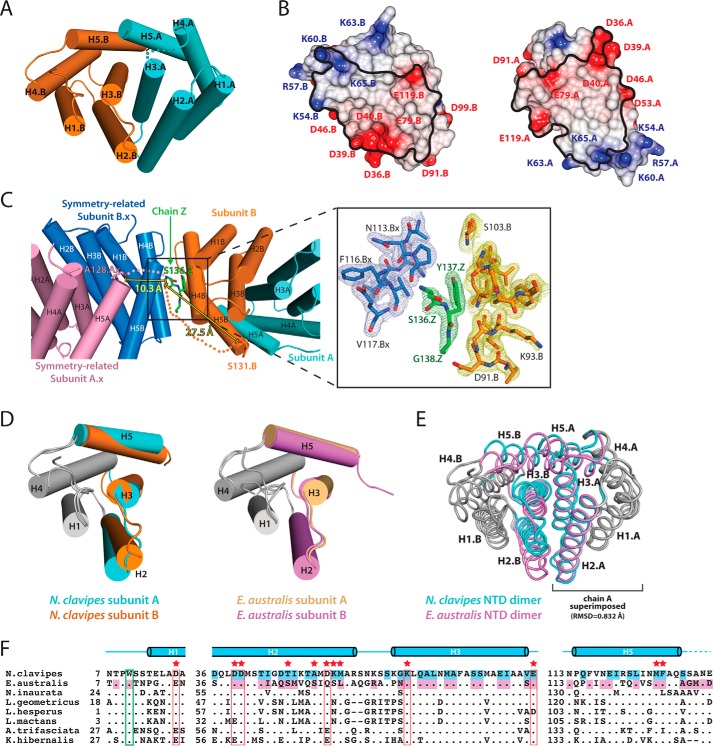
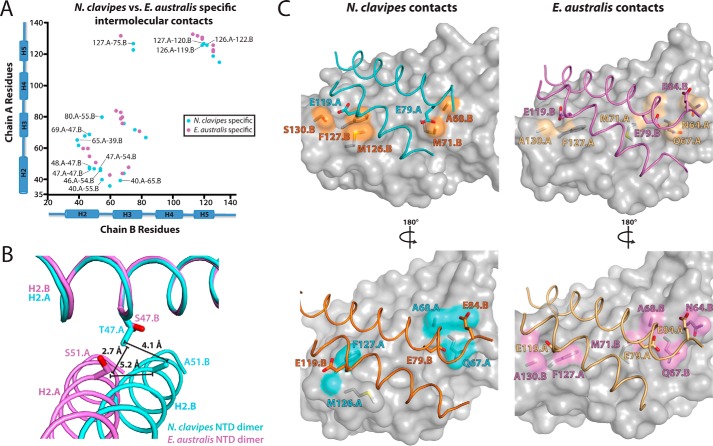
Similar to the EaNTD (10, 11, 14), the NcNTD adopts a five-helix bundle structure, and the two NTD molecules in the crystallographic asymmetric unit form a homodimer in which helices 2, 3, and 5 are involved in all of the contacts across the dimer interface (Fig. 1A). The total buried surface area at the interface of the NcNTD dimer is 2363 Å2, which is comparable with the 2800 Å2 buried at the EaNTD dimer interface. Also similar to the EaNTD structure, there are clusters of basic (Lys54, Arg57, Lys60, Lys64, Lys65) and acidic (Asp36, Asp39, Asp40, Glu79, Asp91) residues at opposite ends of each monomer subunit (Fig. 1B). These charged areas create a dipole moment within each subunit, and the subunits are arranged antiparallel to each other to accommodate the charged poles. The residues responsible for this non-uniform charge arrangement are highly conserved among spidroin NTDs (11) (Fig. 1F).
FIGURE 1.
N. clavipes major ampullate spidroin NTD structure. A, the crystal structure of the NcNTD is shown as a ribbon representation with subunit A colored cyan and subunit B colored orange. Secondary structure elements are labeled H1–H5, and the letters A and B indicate the subunit. The semitransparent cyan spheres represent the disordered loop containing residues 83–86 of subunit A. B, the electrostatic surface potential of the NcNTD shows clusters of positive and negative charges at opposite poles of each subunit. The structure is depicted in open book representation. The black outlines on the surface indicate the regions of each subunit that are making intermolecular contacts. The electrostatic potentials were calculated using the Poisson-Boltzmann method as implemented in CCP4mg with red and blue representing negative and positive electrostatic potential, respectively, on a scale ranging from +0.5 V to −0.5 V. C, chain Z (green) was built as a Ser-Tyr-Gly tripeptide into an area of electron density between subunit B of the NcNTD (orange) in the asymmetric unit and subunit B.x of a symmetry-related NTD molecule (blue). This tripeptide corresponds to C-terminal residues 136–138 of the NcNTD. The Ser-Tyr-Gly tripeptide appears to derive from subunit A of a symmetry-related NTD dimer (pink) based on the distances between the last ordered residue at the C termini of subunits surrounding the tripeptide, and the N terminus of the tripeptide (Ser136). Semitransparent pink and orange spheres represent the potential disordered region connecting the last ordered residues of subunits that are in proximity to the Ser-Tyr-Gly tripeptide. Electron density maps for chain Z (green) and the neighboring residues of subunit B.x (blue) and subunit B (yellow) are shown in the right panel contoured at 1.5σ. D, the subunits of the NcNTD (left) and EaNTD (right) structures were superimposed to highlight the topology of the helices comprising the five-helix bundles. Note that H2, H3, and H5 are oriented differently from H1 and H4 across the two subunits of the NcNTD dimer, whereas the two subunits of the EaNTD structure are virtually identical. The helices that comprise the dimer interface are colored, while non-interacting helices are grayed. E, comparison of the overall NcNTD and EaNTD (PDB: 3LR2) (11) structures. Subunit A of the NcNTD was superimposed with subunit B of the EaNTD as these were the subunits that were most similar to each other. Note the difference in the topology of the helices at the dimer interfaces of the structures. F, the sequence alignment of MaSp NTDs is condensed to show the regions containing key residues involved in NTD dimerization. The secondary structure is shown above the sequence, and the dashed line at the C terminus represents the disordered region. Cyan-shaded residues make intermolecular contacts in the NcNTD structure. Violet-shaded residues make intermolecular contacts in the EaNTD structure. A period indicates a strictly conserved residue, and a dash indicates a gap in the sequence. Residue numbers are shown on the left of each fragment of the alignment. Red boxes indicate residues involved in the key salt bridge and handshake interactions; red stars indicate other key residues at the dimer interface; and the green box highlights the single, highly conserved tryptophan. The sequences chosen for alignment were based on the nucleotide-based distance trees for the major ampullate spidroin genes (8). N. inaurata, Nephila inaurata; L. geometricus, Latrodectus geometricus; L. hesperus, Latrodectus hesperus; L. mactans, Latrodectus mactans; A. trifasciata, Argiope trifasciata; K. hibernalis, Kukulcania hibernalis.
In addition to chains A and B, which form the canonical dimer described above, the NcNTD structure also contains chain Z, which consists of the three amino acids (Ser-Tyr-Gly) that correspond to residues 136–138 of the NcNTD. Although it is unclear as to which chain these residues belong due to varying degrees of disorder at the C termini of the NTD subunits, it appears to derive from a symmetry-related NTD molecule in the crystal (Fig. 1C). The ordering of this Ser-Tyr-Gly tripeptide and the contacts it engages in are most likely an artifact of crystallization.
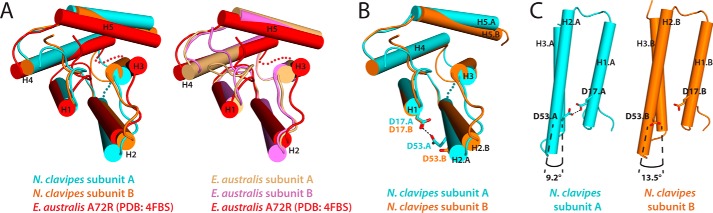
Although the NcNTD and EaNTD structures display general similarities, our structural analysis has revealed key differences that provide new mechanistic insights into pH-dependent NTD dimer formation. Although the subunits of the EaNTD dimer are nearly identical (Fig. 1D, rmsd = 0.505 Å), superposition of the two subunits of the NcNTD dimer (Fig. 1D, rmsd = 0.914 Å) reveals that the arrangements of the helices comprising the five-helix bundles are distinct from each other. Specifically, helices 2, 3, and 5 are oriented differently from helices 1 and 4, which behave as a rigid body (Fig. 1D). Because helices 2, 3, and 5 are involved in all of the intermolecular contacts in the NTD dimer, this topological difference alters how the subunits are arranged relative to each other at the dimer interface (Fig. 1E) and results in a novel network of intermolecular contacts (supplemental Table S1). The details of the differences in subunit topology and the novel contacts that result will be discussed in greater detail below. This is not the first instance where altered helix topology has been observed in the MaSp NTD. The crystal structure for the monomeric EaNTD A72R mutant (PDB: 4FBS) (10) shows distinct topological differences when compared with the subunits of the wild-type EaNTD dimer (see Fig. 3A), and structural rearrangement of the helices comprising the five-helix bundle has been shown to be involved in the monomer to dimer transition (10). These findings, together with the fact that there are few crystal contacts around the dimer interface in the NcNTD structure, suggest that the differences in helix topology and the resulting intermolecular contacts are not due to crystal packing. The differences we observe in our structure have important implications for our understanding of the mechanisms that control NTD dimerization and will be discussed in greater detail below.
FIGURE 3.
Altered helix topologies of monomer and dimer subunits. A, left, the subunits of the NcNTD dimer structure are superimposed with the EaNTD monomer crystal structure (A72R; PDB: 4FBS) (10). Right, the subunits of the EaNTD dimer crystal structure are superimposed with the EaNTD monomer crystal structure. Both superimpositions illustrate the structural rearrangement of the five-helix bundle that occurs during the monomer to dimer transition. The semitransparent cyan and red spheres represent disordered loops in the NcNTD subunit A and EaNTD A72R structures, respectively. B, the two subunits of the NcNTD dimer are shown as a ribbon representation and are superimposed as in Fig. 1D. Side chains for residues Asp17 and Asp53 are shown as sticks, and the handshake interaction (black dashed line) between these two residues in subunit A (cyan) is shown. The semitransparent cyan spheres represent the disordered loop containing residues 83–86 of subunit A. C, H1, H2, and H3 of both NcNTD dimer subunits are shown in a ribbon representation, and residues Asp17 and Asp53 of both subunits are shown as sticks. The subunits are oriented to show the difference in the angle at which H2 crosses H3 due to the presence or absence of the Asp17-Asp53 handshake interaction (black dashed line).
Distinct Asymmetry among Key Salt Bridge and Handshake Interactions
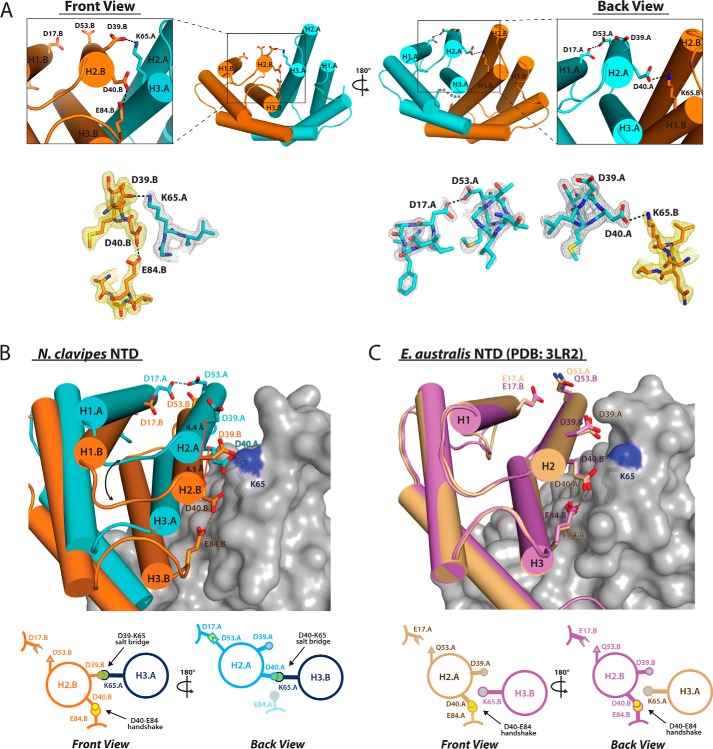
Among the residues that previous structural studies on EaNTD have identified as being particularly important for pH-dependent NTD dimerization are Asp40, Lys65, and Glu84 (11, 13). In the EaNTD crystal structure, the side chains of Asp40 and Glu84 are in very close proximity to each other and form an intramolecular handshake interaction (11) that stabilizes the otherwise unfavorable clustering of two negatively charged residues (15). Although Asp40 and Lys65 do not engage in short-range intermolecular contacts (<3.5 Å), the side chains are within ∼4–4.3 Å, which permits long-range intermolecular electrostatic interactions across the dimer interface. Importantly, the contacts in which Asp40, Lys65, and Glu84 engage are highly symmetrical, meaning that the equivalent residues on both subunits engage in equivalent intra- and intermolecular contacts (Fig. 2C). Mutation of each of these residues leads to a decrease in dimer formation, confirming their involvement in dimerization (12, 13, 16).
FIGURE 2.
Comparison of key intra- and intermolecular interactions in the NcNTD and EaNTD structures. A, top, front and back views of the NcNTD dimer reveals differences in the pattern of key salt bridge and handshake interactions taking place between equivalent residues on the two dimer subunits with subunit A colored cyan and subunit B colored orange. Bottom, 2.02 Å 2Fo − Fc electron density maps (contoured at 1.5σ) for key salt bridge and hand shake interactions are shown in the same orientation as the top panel. B, top, to illustrate the asymmetry of the NcNTD dimer interface, two copies of the NTD dimer were overlaid by superimposing the rigid body of subunit A of one dimer with the rigid body of subunit B of the other. The subunits superimpose well with an rmsd of 0.525 Å, and for clarity, only one copy is shown as a surface representation with the side chain of Lys65 shown as sticks. The other subunits of the dimers are shown as ribbons with residues involved in key intra- and intermolecular contacts shown as sticks. For a highly symmetric dimer there would be two sets of equivalent interactions. Bottom, graphic illustrating key intra- and intermolecular interactions at the NcNTD dimer interface. C, top, two copies of the EaNTD dimer (PDB: 3LR2) (11) were superimposed and are shown as described in panel B. The EaNTD dimer is more symmetric when compared with the NcNTD dimer, and this is reflected by two sets of equivalent intra- and intermolecular interactions taking place between the subunits. Bottom, graphic illustrating key intra- and intermolecular interactions at the EaNTD dimer interface.
As a result of the altered subunit topology at the interface observed in the NcNTD structure, the interactions that Asp40, Lys65, and Glu84 are involved in are surprisingly different from those of the EaNTD. On one side of the dimer, which we refer to as the “front” side for clarity, Asp40 and Glu84 engage in the intramolecular handshake interaction as observed in the E. australis structure (Fig. 2A, top row, left). However, rather than only engaging in long-range electrostatic interactions, Lys65 engages in a short-range (2.6 Å) intermolecular salt bridge with Asp39. On the other side of the dimer, which we refer to as the “back” side for clarity, rather than engage in the intramolecular handshake interaction with Glu84 (which is disordered in this subunit), Asp40 engages in a short-range (3.1 Å) intermolecular salt bridge with Lys65 (Fig. 2A, top row, right). Asp39 is not involved in contacts on the back side of the dimer. The electron density maps for all of the residues involved in these contacts were of excellent quality (Fig. 2A, bottom row).
These contacts differ from the EaNTD crystal structure due to both their asymmetric nature and the involvement of Asp39. The role of Asp39 in NTD dimerization was not initially investigated, likely due to the fact that its side chain is 3.5–5 Å away from any basic side chain atoms on the adjacent subunit in the EaNTD crystal structure and is not within hydrogen-bonding distance in the majority of NMR conformers reported for the NTD dimer. However, a recent study investigating the effects of systematic mutation of each of the eight acidic residues in the EaNTD on dimerization revealed that a D39N substitution almost completely abolishes dimer formation (16). Thus, our structure provides a molecular basis for the critical role of Asp39 in NTD dimerization as it clearly demonstrates that this residue engages in short-range intermolecular contacts to Lys65 across the dimer interface (Fig. 2A).
The nature of the contacts that take place between Asp39, Asp40, and Lys65 in the NcNTD and EaNTD structures is reflective of the topology of the subunits at the dimer interfaces. In a highly symmetric dimer as observed in the EaNTD structure, Asp39 and Asp40 of both subunits engage in equivalent contacts at both ends of the dimer interface (Fig. 2C). The asymmetric nature of the NcNTD dimer results in a 4.4 and 4.5 Å translation of Asp39 and Asp40 with respect to Lys65 on the adjacent subunit, which allows formation of the Asp39-Lys65 salt bridge at one end of the dimer and the Asp40-Lys65 salt bridge at the other (Fig. 2B). Despite these differences in subunit topology, the asymmetric Asp39-Lys65 and Asp40-Lys65 salt bridges in the NcNTD perform a similar structural role as the symmetric long-range electrostatic interactions between these residues in the EaNTD, which is to facilitate proper subunit alignment in the dimer.
Consecutive conserved acidic residues at positions 39 and 40 allow for variability in the contacts that take place to Lys65 in the context of the moderately differing dimer topologies observed in the NcNTD and EaNTD structures. This provides a mechanism for plasticity in the intermolecular contacts at the dimer interface that could be relevant during the transition from loosely to stably associated dimer. The fact that mutation of Asp39 abolishes NTD dimerization is consistent with the idea that structural plasticity plays a role in dimerization, as Asp39 engages in a key short-range salt bridge in our NcNTD structure but not in the more symmetric EaNTD structure (13, 16).
A Novel Asp17-Asp53 Intramolecular Handshake Interaction in the N. clavipes NTD
During our structural analysis, we observed a novel intramolecular handshake interaction that takes place between Asp17 and Asp53 in one subunit of the NcNTD (Fig. 2A). Asp17 and Asp53 are highly conserved as acidic residues across spider species; however, this intramolecular handshake interaction was not observed in the EaNTD structure because it is an outlier harboring a glutamine at position 53 (Fig. 1F). One of the key differences across the subunits is that H2 tilts significantly toward H1 in the subunit in which this handshake interaction between Asp17 and Asp53 takes place (Fig. 3, B and C). This suggests that the asymmetric nature of the Asp17-Asp53 handshake interaction partially accounts for the differing five-helix bundle topologies of the NcNTD dimer subunits described above (Fig. 1D), and by extension, the asymmetric intermolecular interactions observed at the dimer interface (supplemental Table S1).
Glutamate 84, which engages in the Asp40-Glu84 handshake interaction, is known to be a key player in the pH-dependent relay mechanism that controls NTD dimerization (13). The proximity of negatively charged carboxylates can lead to a substantial elevation of side chain pKa to a physiological value. The pH decrease experienced by the NTD as it progresses through the spinning duct leads to protonation of these groups, which serves to alleviate electrostatic repulsion. It is through such a mechanism that protonation of Glu84 is proposed to facilitate a structural transition that leads to formation of the stably associated dimer. Given the aforementioned link between the topology of the helices in the five-helix bundles of the NTD subunits and the presence or absence of the novel Asp17-Asp53 handshake interaction (Fig. 3, B and C), as well as the similar chemical environments experienced by the Asp17-Asp53 and Asp40-Glu84 handshakes (Fig. 2B), it is tempting to speculate that either Asp17 and Asp53 are an additional part of the pH-sensitive relay mechanism for dimerization in spider species in which the two acidic residues are present. Indeed, an additional level of complexity in the pH-sensitive relay mechanism has recently been demonstrated in a study of Araneus ventricosus minor ampullate spidroin, which lacks an acidic residue at position 84, but instead has a titratable non-conserved acidic residue (Glu73) at the position corresponding to Ser76 of the N. clavipes NTD (17). We further investigate the role that the Asp17 and Asp53 residues play in dimerization below.
Altered Arrangement of NTD Dimer Subunits Results in Unique Intermolecular Contacts
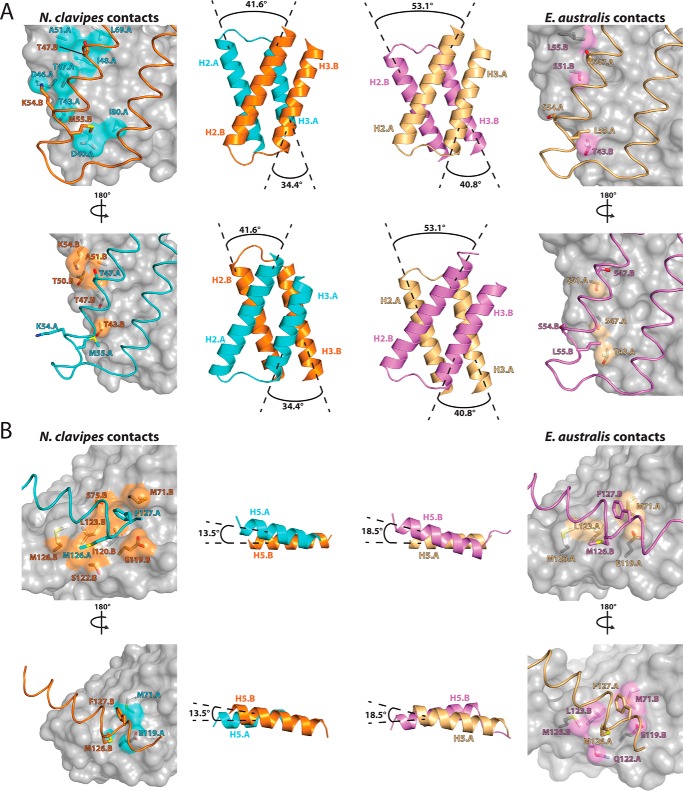
Although the residues involved in the asymmetric salt bridge and handshake interactions described above play a well defined role in dimer formation, there are a number of less-studied residues at the NcNTD dimer interface that engage in novel asymmetric contacts (supplemental Table S1). The majority of the novel contacts observed in our structure result from the distinct arrangement of the three helices (H2, H3, and H5) involved in all of the intermolecular contacts at the dimer interface (Fig. 1D), rather than differences in protein sequence. This distinct helix arrangement results in a significant increase in asymmetric contacts (supplemental Table S2) at the NcNTD dimer interface (60%; 30 of 50 total contacts) when compared with the EaNTD (39%; 20 of 52 total contacts).
To illustrate the differences in how the interfacial helices are arranged at the dimer interface, we calculated the angles at which helices 2, 3, and 5 cross each other in the NcNTD and EaNTD structures. This analysis shows that the angles at which helices 2, 3, and 5 cross their counterparts in the N. clavipes and E. australis NTD structures differs by 11.5, 6.4, and 5.0°, respectively (Fig. 4, A and B, middle panels). As a result of the altered helix topologies, the percentage of asymmetric contacts involving residues on helices 2 and 3, which account for most of the interactions at the interface, increases from 32% (12 of 37) for EaNTD to 62.5% for NcNTD (25 of 40). Likewise, the altered positioning of H5 increases asymmetric contacts engaged in by residues on this helix from 56% (10 of 18) in the EaNTD to 64% (9 of 14) in the NcNTD (supplemental Table S2).
FIGURE 4.
Differences in the topology of the subunits at the NTD dimer interface results in unique intermolecular contacts. A, top, in the left (NcNTD) and right (EaNTD) panels, one NTD subunit of each dimer is shown as a semitransparent gray surface, and the other is shown as ribbons for the H2/H3 dimer interface. Side chains of residues that engage in novel contacts resulting from the altered interfacial helix topologies are shown as sticks, and the surface is colored as in the center panel. Bottom, to allow for comparison of contacts at the opposite side of the NTD dimer interface, the structures were rotated 180° about the y axis and are presented in the same manner as in the top row. Middle, to illustrate the differences in how the interfacial helices are arranged at the dimer interface, we calculated the angles at which H2/H3 cross each other in the NcNTD and EaNTD structures, which are shown in the same orientation as ribbons. Angles were calculated using the program Chimera. B, unique intermolecular contacts at the H5 dimer interface of the NcNTD (left) and EaNTD (right) are presented in the same manner as in (A). (middle) The angle at which H5.A crosses H5.B in each NTD structure is shown.
The topologically distinct manner in which the interfacial helices of the NcNTD and EaNTD are related to each other at the dimer interface results in novel networks of intermolecular contacts. Notably, residues T47.B, K54.B, and M55.B3 on H2B of the NcNTD are significantly more buried at the dimer interface (Fig. 4A, top row, left panel) than the corresponding residues of the EaNTD (Fig. 4A, top row, right panel). On one side of the NcNTD, T47.B and M55.B on H2 engage in a unique network of van der Waals contacts to D40.A, T43.A, I48.A, A51.A, and L69.A on helices 2 and 3 of the adjacent subunit. K54.B of H2B also engages in a unique hydrogen bond to T43.A and electrostatic interaction with D46.A on H2A. Almost all of these contacts rely on the distinct way in which H2 and H3 sit across from each other at the NcNTD dimer interface in order for the residues to be properly positioned for intermolecular interactions. The equivalent residues on the opposite side of the NcNTD dimer interface also engage in specific, albeit distinct sets of contacts as a result of the altered topology of the interfacial helices and further highlight the increased asymmetry of the NcNTD intermolecular contacts when compared with those of the EaNTD. In the NcNTD, T47.A, K54.A, and M55.A are significantly less buried than their counterparts in subunit B, especially K54.A, which does not engage in any intermolecular contacts (Fig. 4A, bottom row, left panel). By comparison, S47.B, S54.B, and L55.B of the EaNTD form three intermolecular contacts combined (Fig. 4A, bottom row, right panel), mirroring their counterparts in subunit A.
With respect to H5, a network of unique H5-H3 and H5-H5 interactions further highlights the role that altered interfacial helix topology plays in determining the plastic nature of the NTD dimer interface. Notably, M126.A and F127.A from H5 of the NcNTD (Fig. 4B, top row, left panel) are significantly more buried at the dimer interface relative to the corresponding EaNTD residues (Fig. 4B, top row, right panel). Specifically, in the NcNTD structure, F127.A inserts into a hydrophobic pocket formed by the side chains of M71.B, S75.B, E119.B, and I120.B on helices 3 and 5, and M126.A sits on a hydrophobic patch formed by S122.B, L123.B, and M126.B on H5. Interestingly, the hydrophobic pocket into which F127.A inserts in the NcNTD structure is not present in the EaNTD structure due to the pocket being filled intramolecularly by the side chain of M126.A. As a result, if the interfacial helices of the EaNTD dimer interface adopted a topology similar to the NcNTD, Phe127 would engage in severe steric clashes with residues across the dimer interface. Similar to the above observations for the H2 and H3 interactions, the H5 interactions also display distinct differences in symmetry between the NTD orthologs. In the NcNTD, M126.B and F127.B engage in significantly fewer intermolecular contacts than their counterparts in subunit A (Fig. 4B, bottom row, left panel), whereas the intermolecular contacts involving Met126 and Phe127 in the EaNTD are completely symmetric (Fig. 4B, bottom row, right panel).
Overall, the NcNTD contains clusters of contacts across the dimer interface not seen in the EaNTD, including unique H2-H2 contacts, H2-H3 contacts, and H5-H5 contacts (Fig. 5A). Among the group of aforementioned H2-H2 contacts, the intermolecular interactions between residues 47 and 51, which are almost completely buried at the dimer interface of the two NTD orthologs, are of particular interest. In the NcNTD, Thr47 engages in symmetric van der Waals contacts with Ala51 (Fig. 5B), whereas in the EaNTD, Ser47 and Ser51 both adopt alternative conformations in the crystal structure that facilitate Ser51-Ser51 or Ser51-Ser47 hydrogen bonds (supplemental Table S1). Thr47 and Ala51 are both strictly conserved in most MaSp1 orthologs (Fig. 1F) with E. australis being an exception. The angle at which H2A crosses H2B in the NcNTD increases the distance between the distal atoms of the Thr47 and Ala51 side chains by about 1.5 Å relative to the EaNTD, and this subtle shift creates space that is required to accommodate the larger Thr47 side chain (Fig. 5B). Due to the closer proximity of residues 47 and 51 in the EaNTD structure, the β branched Thr47 side chain present in most other MaSp1 orthologs could not be accommodated at position 47 regardless of the side chain rotamers adopted by the interacting residues. These observations suggest that relatively subtle amino acid substitutions could unexpectedly contribute to structural plasticity at the NTD dimer interfaces of different MaSp1 orthologs, similar to what we observe in the EaNTD and NcNTD structures.
FIGURE 5.
Equivalent intermolecular contacts at NcNTD and EaNTD dimer interfaces. A, a protein contact map shows where the unique intermolecular contacts are clustered on the secondary structure elements of the NcNTD (cyan) and EaNTD (violet) structures. The protein contact maps are restricted to show only unique intermolecular contacts. Residue numbers are indicated on the x and y axes. The secondary structure of the subunits is also shown on the axes to facilitate identification of where the unique contacts are occurring in the context of the structure. Unique contacts that were discussed under “Results and Discussion” are indicated with lines, and the residue numbers are shown. B, the NcNTD and EaNTD structures were superimposed and are depicted as described in Fig. 1D. The β branched side chain of NcNTD T47.A, which is conserved in most MaSp1A orthologs but is a serine in EaNTD, can be accommodated as a result of the difference in the topology of the interfacial helices, in particular H2. Distances between atoms discussed under “Results and Discussion” are indicated. C, top, H3 and H5 of one subunit of the N. clavipes (left) and E. australis (right) NTD dimer are shown as backbone worm representations with the side chains for conserved acidic residues Glu79, Glu84, and Glu119 shown as sticks. The other subunits of the dimers are shown as semitransparent gray surface representations, with residues involved in contacts to the conserved acidic residues shown as sticks and their surfaces colored as in Fig. 4A. Bottom, to allow for comparison of contacts on both sides of the NTD dimer interface, the structures were rotated 180° about the y axis and are presented in the same manner as in the top row.
Interestingly, the intermolecular contacts for the highly conserved acidic residues Glu79, Glu84, and Glu119 are more asymmetric in the NcNTD (3 of 6) than in the EaNTD (1 of 6) (Fig. 5C). These residues have been shown to be critical to the pH-sensitive relay mechanism in the EaNTD dimerization process due to their involvement in the formation of the weakly associated dimer (13). Thus, the differences in symmetry observed for the contacts these residues are involved in may play a mechanistic role in pH-dependent NTD dimerization.
Novel Intra- and Intermolecular Interactions Are Involved in NcNTD Dimerization
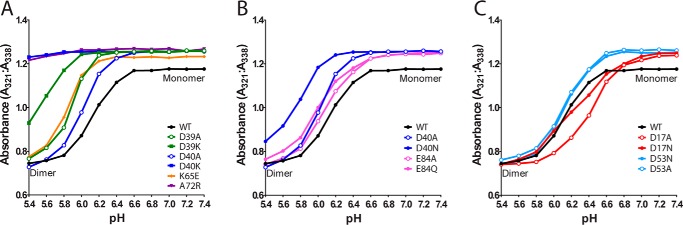
To investigate the role that residues involved in the novel asymmetric intra- and intermolecular interactions observed in the NcNTD structure play in pH-dependent NTD dimerization, we made use of a tryptophan fluorescence assay that was developed and has routinely been used by groups studying spidroin NTDs (7, 11, 13, 17). Similar to the EaNTD, the NcNTD contains a single conserved tryptophan near the N terminus (Trp10) (Fig. 1F). During transition of the NTD from monomer to dimer, Trp10 undergoes a conformational change that increases its solvent exposure and results in quenching of its intrinsic fluorescence emission (7, 10, 13). To probe the pH-dependent dimerization of the wild-type and mutant NcNTD, we excited the proteins at 280 nm and recorded their emission spectra between 300–400 nm from a pH range of 7.4 to 5.4. The ratio between the fluorescence intensity at 321–338 nm at each pH step represents the ratio of the NcNTD in its monomeric and dimeric states, respectively (supplemental Fig. S1), similar to the case for the EaNTD. The WT NcNTD behaved as expected (7), with the monomer to dimer transition point at ∼pH 6.1 (Fig. 6A). Also as expected, the A72R mutant, which was designed to introduce a sterically and electrostatically unfavorable side chain into the center of the dimer interface (10), retained a high 321:338 ratio throughout the pH range studied, indicating that it served as an adequate monomer control.
FIGURE 6.
Structure-function analysis of residues involved in NcNTD dimerization. During transition of the NTD from monomer to dimer, the single tryptophan residue of the NTD (Trp10) undergoes a conformational change that increases its solvent exposure and results in quenching of its intrinsic fluorescence emission. The ratio of the tryptophan fluorescence signals at 321–338 nm reflects the ratio between monomeric (338 nm) and dimeric (321 nm) NcNTD. A–C, the tryptophan fluorescence ratio of the signals at 321 and 338 nm was plotted as a function of pH for WT, A72R, and mutants intended to affect the Asp39-Lys65 and Asp40-Lys65 salt bridges (A), mutants probing the Asp40-Glu84 handshake interaction (B), and mutants probing the Asp17-Asp53 handshake interaction (C). Experiments were performed in triplicate, and representative raw data are shown in supplemental Fig. S1.
The tryptophan fluorescence data for mutations of residues involved in the asymmetric Asp40-Lys65 and Asp39-Lys65 salt bridges observed in our structure confirms the critical role of these residues in pH-dependent NTD dimerization. Similar to results seen for the EaNTD (13), disruption of the Asp40-Lys65 salt bridge via the point mutants D40A, D40K, and K65E results in an apparent decrease in dimer stability (Fig. 6A), with the D40K mutation completely abolishing dimer formation in both NTD orthologs. As mentioned above, Asp39 does not engage in short-range intermolecular contacts in the EaNTD crystal structure and the majority of NMR conformers. This is likely the reason that the critical role that Asp39 plays in NTD dimerization was not discovered until a recent study in which all of the acidic residues in the EaNTD were indiscriminately mutated (16). The results of our tryptophan fluorescence studies probing D39A and D39K NcNTD mutations confirm the importance of Asp39 in pH-dependent NTD dimerization, and our crystal structure provides a molecular basis for the role that Asp39 plays in this process due to its short-range intermolecular salt bridge to Lys65. Together, our data indicate that Asp39, Asp40, and Lys65 are essential to the NcNTD dimerization process, although we cannot distinguish between a role for the short-range asymmetric salt bridges observed in the NcNTD structure versus the symmetric long-range electrostatic interactions observed in the EaNTD structure.
The asymmetric Asp40-Glu84 intramolecular handshake interaction had not been previously investigated in the NcNTD. Thus, we generated an E84A mutant designed to completely disrupt the handshake and an E84Q mutant designed to mimic the protonated state of glutamate that is posited to be involved in dimer stabilization as the spidroin progresses through the spinning duct. Indeed, our results show that the E84A substitution destabilizes dimer formation, underscoring the importance of the handshake in this process (Fig. 6B). The E84Q mutant is also slightly dimer-destabilizing, which is consistent with the results of the corresponding single point mutation in EaNTD. Kronqvist et al. (13) showed that the stabilizing effects that protonation of Glu84 has on NTD dimerization must be preceded by protonation of Glu79 and Glu119. Thus, altering the charge of this individual residue does not significantly affect the monomer-dimer equilibrium. Together, our data are consistent with the idea that NcNTD residues Glu79, Glu84, and Glu119 are part of a pH relay system similar to that previously observed in the EaNTD (Fig. 5C) (13).
The most surprising results involve the mutations designed to affect the novel intramolecular Asp17-Asp53 handshake interaction observed in our NcNTD structure (D17A, D17N, D53A, and D53N), as shown in Fig. 6C. D17N, D53A, and D53N did not significantly affect the dimer stability when compared with WT, whereas D17A facilitated dimer formation at a higher pH when compared with WT. These data imply that affecting the charge at residue 17 has a significant effect on dimer stability, but the mechanisms that explain this effect are unclear. It is a formal possibility that protonation of either Asp17 or Asp53 must be accompanied by the protonation of other acidic residues, similar to the mechanism for Glu84 protonation described above, and this will be the focus of future investigations.
Conclusions
In this study, we have reported the 2.02 Å crystal structure of the N. clavipes MaSp1 NTD dimer. Notably, the topology of the NTD subunits at the dimer interface of our structure is distinct from that observed in the previously reported crystal and NMR structures of the E. australis NTD (11, 13). Although there is an overlapping network of conserved interactions at the dimer interface of both orthologs, this topological difference results in a subset of contacts that are specific to each ortholog, as well as substantially more asymmetry in the interactions observed at the NcNTD dimer interface. Our structural and biochemical findings underscore the importance of residues Asp39, Asp40, and Lys65 to pH-dependent NTD dimerization. The presence of consecutive acidic residues that are fully conserved at positions 39 and 40 of NTDs allows for plasticity in the interactions these residues engage in depending on precisely how the subunits are arranged at the dimer interface. This results in asymmetric Asp39-Lys65 and Asp40-Lys65 salt bridges in our structure versus symmetric long-range electrostatic interactions between these residues in the EaNTD. These sets of contacts play structurally equivalent roles in aligning the subunits to each other. We also identify a novel intramolecular handshake between conserved acidic residues at positions 17 and 53 that we show plays an important role in the pH-dependent dimerization of the NcNTD. Our data suggest that these residues may participate in the pH relay mechanism established by Kronqvist et al. (13), although further studies will be required to test this possibility.
It has been established that a rearrangement of the five-helix bundle of the NTD subunit is necessary for NTD dimer formation (10), and a “conformational selection” model (18) for NTD dimerization has been proposed in which the five-helix bundle of NTD subunits populates many conformations that are in dynamic equilibrium. Dimerization occurs when an NTD subunit selects a partner with the complementary binding interface from an ensemble of conformers. Our structure is the first to show two significantly different subunit topologies in one dimer, and the plasticity seen in the NcNTD dimer interface could be a contributing factor to the conformational selection process that regulates the monomer to dimer transition and/or the transition from loosely to stably associated dimer. Thus, the NcNTD structure presented here and the previously published MaSp NTD structures (10, 11, 13) could be on pathway snapshots of the NTD during the pH-dependent dimerization process that occurs as the spidroins progress through the spinning duct.
Finally, our study reveals new insights on the molecular evolution of different types of spider silk. Although our work and the work of others (11, 13, 17) have highlighted subtle differences in the pH relay system and structural plasticity at the dimer interface of major and minor ampullate spidroin NTDs across various species, the overall mechanism of pH and salt-dependent NTD dimerization is likely to be highly conserved. In contrast, tubuliform spidroins, which are a more evolutionarily ancient spidroin used for egg case construction in virtually all spiders, lack many of the key residues involved in the pH relay system that has been studied in EaNTD and NcNTD, including Asp17, Asp39, Asp53, and Glu119 (19). Although major and minor ampullate spidroins must be spun in a very rapid manner by spiders for web construction and their lifeline on a daily basis, tubuliform spidroins are only used a few times in a female spider's lifetime to create a protective casing around eggs. We speculate that the poor conservation of these key acidic residues in tubuliform spidroin NTDs may reflect a lack of evolutionary pressure to very rapidly spin this type of silk, whereas the presence of the pH relay mechanism has evolved to make this possible for major and minor ampullate spidroins. Further, the fact that acidic residue Asp39, which has been shown to be essential for NTD dimerization in major ampullate spidroins (16), is not conserved in tubuliform spidroins suggests that there may be additional differences in the mechanisms of NTD dimerization across different spidroin types, although this would require additional experiments to confirm.
Experimental Procedures
Protein Expression and Purification
All proteins in this study were expressed and purified using a slight variation of a previously published protocol (7). Briefly, the proteins were expressed by transforming the expression plasmids into BL21-DE3 (Stratagene) cells via heat-shock transformation. Large-scale cultures were grown in an incubated shaker at 37 °C to an A600 of 0.8. Protein overexpression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside, and the cultures were shaken an additional 18 h at 18 °C. Cells were harvested via centrifugation, and the pellets were re-suspended in buffer containing 20 mm Tris, pH 8.0, with 350 mm NaCl. The cells were lysed by sonication and purified using a GSTrap 4B column or GSTrap agarose gel bed (GE Healthcare Life Sciences). Affinity-tagged protein was eluted using a buffer containing 15 mm glutathione in 50 mm Tris, pH 8.0, 1 mm DTT. The samples were then buffer-exchanged to the same buffer used for re-suspension of the pellets. The GST tag was cleaved by incubating the purified fusion protein with PreScission Protease (GE Healthcare Life Sciences) at a ratio of 1:1000 (w/w) overnight at 4 °C. After GST removal, 10 vector-derived residues (GPLGSPGIPG) remained on the N-terminal end of the mature protein. The samples were then subjected to gel filtration (HiLoad 26/600 Superdex 75 pg, Amersham Biosciences) and ion exchange (MonoQ 10/100 GL, Amersham Biosciences) to separate the NcNTD from the GST tag. The purity of each purified sample was assessed by SDS-PAGE using either a 15% or 17% acrylamide gel stained with Coomassie Brilliant Blue. The pure NTD was then concentrated and/or buffer-exchanged as needed. All point mutations were introduced using PCR-based mutagenesis and expressed and purified using the same protocol as for wild type.
X-ray Crystallography
Wild-type NcNTD was purified as described and concentrated to a final concentration of 10.1 mg/ml in a buffer containing 20 mm Tris, pH 8.0, and 50 mm NaCl. The Index (Hampton Research) and JCSG Core I, II, III, and IV (Qiagen) commercial screens were used for sparse-matrix screening in 96-well Greiner microplates (400-nl sitting drop vapor diffusion format) to identify conditions suitable for crystal growth. The Crystal Gryphon robot (Art Robbins Instruments) was used to set these trays. Diffraction quality crystals were grown by manually mixing 1.0 μl of protein with 1.0 μl of well solution (0.1 m Bis-Tris, pH 6.5, 29% w/v PEG 3350) by hanging drop vapor diffusion at 18 °C. Crystals formed within 1–2 days and were harvested by flash-freezing in liquid nitrogen in mother liquor plus 10% ethylene glycol as a cryoprotectant. The protein crystallized in the P3121 space group with unit cell dimensions a = 67.48, b = 67.48, and c = 90.35. There was one copy of the NTD dimer in the asymmetric unit, and the crystal had an estimated solvent content of 38.0%. Diffraction data sets were collected on the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. All data were indexed, integrated, and scaled using HKL2000 (20).
Structure Determination and Refinement
A data set was collected to a resolution of 2.02 Å for the N. clavipes major ampullate spidroin NTD. The Rmerge values for the two highest resolution shells of this data set were 46 and 60%, respectively. We included the highest resolution shell due to its high I/σ (5.6) and CC½ (93.5) (where CC½ is correlation coefficient) and based on improved electron density map quality when compared with alternatively processed data with lower resolution cut-offs. The structure of the E. australis major ampullate spidroin NTD (PDB: 3LR2) (11) was used as the model for molecular replacement using the PHASER software (21). The model was refined to Rcryst/Rfree values of 0.164/0.205 by iterative refinement using PHENIX (22) and COOT (23) software. The final N. clavipes MaSp1A NTD model contains residues 6–82 and 87–128 in subunit A and residues 6–131 in subunit B. Chain Z contains residues 136–138, which appear to derive from subunit A of a symmetry-related molecule (Fig. 1C). The refined structure also contained 123 water molecules. The detailed data collection and refinement statistics are listed in Table 1. Molecular graphics representations were generated using PYMOL (24) and the CCP4mg program of the CCP4 6.4.0 software suite (25).
Tryptophan Fluorescence
Stock buffer solutions were made using a 20 mm MES/20 mm HEPES buffer system to cover the pH range of 5.4–7.4 in 0.2 pH increments. Working buffers were made to contain 154 mm sodium chloride to reflect physiological conditions (26). Each mutant was expressed and purified as described and was buffer-exchanged to a stock solution containing only 50 mm NaCl and then concentrated to 2 mm protein. Five μl of concentrated protein was mixed with 995 μl of each working buffer, so that the final protein concentration was 10 μm. The resulting 1-ml sample was left at room temperature for 30 min to allow the protein to equilibrate. The pH-adjusted samples were transferred to a 1 × 1-cm quartz cuvette and placed in the spectrofluorometer (QuantaMaster 300, Photon Technology International). The samples were excited at 280 nm using a 1-nm bandwidth, and emission spectra were recorded in 1-nm steps between 300 and 400 nm using a 1-nm bandwidth (protocol was adopted from Kronqvist et al. (13)). The ratio between the signals at 321–338 nm, which reflects the ratio between monomeric and dimeric NTD, was calculated for each pH step to compare all mutants across the measured pH range. As a control, the buffer for each pH step was excited without protein added to ensure that the measured signals for the protein samples were not artifacts of the buffer components.
Author Contributions
J. H. A. and S. K. O. designed the study, performed and analyzed all experiments, and wrote the paper. W. R. M. provided conceptual input. S. P. analyzed tryptophan fluorescence data. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-ENG-38. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS from the National Institutes of Health (Grant P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by an National Institutes of Health Office of Research Infrastructure Programs High End Instrumentation grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by the Argonne National Laboratory under Contract DE-AC02-06CH11357. The x-ray crystallography facility used for this work is supported by the Office of the Vice President for Research at the Medical University of South Carolina. The crystallization robotics instrument used in this study was purchased via a National Institutes of Health Shared Instrumentation Award (S10 RR027139-01). We thank Dr. Mirko Hennig and Dr. Daniella Ishimaru for providing insights and advice at early stages of the project. We also thank Katelyn Williams, Dr. Zongyang Lyu, Dr. Lingmin Yuan, and Dr. Christopher Davies for critically reviewing this manuscript.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. S1 and supplemental Tables S1 and S2.
The atomic coordinates and structure factors (code 5IZ2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Throughout the text, in designations such as T47.B, K54.B, and so forth, A or B indicates the subunit of the residue.
- MaSp
- major ampullate spidroin
- NTD
- N-terminal domain
- NcNTD
- N. clavipes major ampullate spidroin 1A NTD
- EaNTD
- E. australis major ampullate spidroin 1 NTD
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- rmsd
- root mean square deviation.
References
- 1. Vollrath F., and Knight D. P. (1999) Structure and function of the silk production pathway in the spider Nephila edulis. Int. J. Biol. Macromol. 24, 243–249 [DOI] [PubMed] [Google Scholar]
- 2. Wendt H., Hillmer A., Reimers K., Kuhbier J. W., Schäfer-Nolte F., Allmeling C., Kasper C., and Vogt P. M. (2011) Artificial skin: culturing of different skin cell lines for generating an artificial skin substitute on cross-weaved spider silk fibres. PLoS ONE 6, e21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allmeling C., Jokuszies A., Reimers K., Kall S., Choi C. Y., Brandes G., Kasper C., Scheper T., Guggenheim M., and Vogt P. M. (2008) Spider silk fibres in artificial nerve constructs promote peripheral nerve regeneration. Cell Prolif. 41, 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gellynck K., Verdonk P. C., Van Nimmen E., Almqvist K. F., Gheysens T., Schoukens G., Van Langenhove L., Kiekens P., Mertens J., and Verbruggen G. (2008) Silkworm and spider silk scaffolds for chondrocyte support. J. Mater. Sci. Mater. Med. 19, 3399–3409 [DOI] [PubMed] [Google Scholar]
- 5. Adrianos S. L., Teulé F., Hinman M. B., Jones J. A., Weber W. S., Yarger J. L., and Lewis R. V. (2013) Nephila clavipes Flagelliform silk-like GGX motifs contribute to extensibility and spacer motifs contribute to strength in synthetic spider silk fibers. Biomacromolecules 14, 1751–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vollrath F. (2000) Strength and structure of spiders' silks. J. Biotechnol. 74, 67–83 [DOI] [PubMed] [Google Scholar]
- 7. Gaines W. A., Sehorn M. G., and Marcotte W. R. (2010) Spidroin N-terminal domain promotes a pH-dependent association of silk proteins during self-assembly. J. Biol. Chem. 285, 40745–40753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaines W. A. 4th, and Marcotte W. R. Jr. (2008) Identification and characterization of multiple Spidroin 1 genes encoding major ampullate silk proteins in Nephila clavipes. Insect Mol. Biol. 17, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vollrath F., and Knight D. P. (2001) Liquid crystalline spinning of spider silk. Nature 410, 541–548 [DOI] [PubMed] [Google Scholar]
- 10. Jaudzems K., Askarieh G., Landreh M., Nordling K., Hedhammar M., Jörnvall H., Rising A., Knight S. D., and Johansson J. (2012) pH-dependent dimerization of spider silk N-terminal domain requires relocation of a wedged tryptophan side chain. J. Mol. Biol. 422, 477–487 [DOI] [PubMed] [Google Scholar]
- 11. Askarieh G., Hedhammar M., Nordling K., Saenz A., Casals C., Rising A., Johansson J., and Knight S. D. (2010) Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465, 236–238 [DOI] [PubMed] [Google Scholar]
- 12. Wallace J. A., and Shen J. K. (2012) Unraveling a trap-and-trigger mechanism in the pH-sensitive self-assembly of spider silk proteins. The J. Phys. Chem. Lett. 3, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kronqvist N., Otikovs M., Chmyrov V., Chen G., Andersson M., Nordling K., Landreh M., Sarr M., Jörnvall H., Wennmalm S., Widengren J., Meng Q., Rising A., Otzen D., Knight S. D., et al. (2014) Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 5, 3254. [DOI] [PubMed] [Google Scholar]
- 14. Hagn F., Thamm C., Scheibel T., and Kessler H. (2011) pH-dependent dimerization and salt-dependent stabilization of the N-terminal domain of spider dragline silk: implications for fiber formation. Angew. Chem. Int. Ed. Engl. 50, 310–313 [DOI] [PubMed] [Google Scholar]
- 15. Flocco M. M., and Mowbray S. L. (1995) Strange bedfellows: interactions between acidic side-chains in proteins. J. Mol. Biol. 254, 96–105 [DOI] [PubMed] [Google Scholar]
- 16. Schwarze S., Zwettler F. U., Johnson C. M., and Neuweiler H. (2013) The N-terminal domains of spider silk proteins assemble ultrafast and protected from charge screening. Nat. Commun. 4, 2815. [DOI] [PubMed] [Google Scholar]
- 17. Otikovs M., Chen G., Nordling K., Landreh M., Meng Q., Jörnvall H., Kronqvist N., Rising A., Johansson J., and Jaudzems K. (2015) Diversified structural basis of a conserved molecular mechanism for pH-dependent dimerization in spider silk N-terminal domains. Chembiochem 16, 1720–1724 [DOI] [PubMed] [Google Scholar]
- 18. Ries J., Schwarze S., Johnson C. M., and Neuweiler H. (2014) Microsecond folding and domain motions of a spider silk protein structural switch. J. Am. Chem. Soc. 136, 17136–17144 [DOI] [PubMed] [Google Scholar]
- 19. Garb J. E., Ayoub N. A., and Hayashi C. Y. (2010) Untangling spider silk evolution with spidroin terminal domains. BMC Evol. Biol. 10, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeLano W. L. (2015) The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 25. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knight D. P., and Vollrath F. (2001) Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften 88, 179–182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.