Abstract
Genetic and genomic studies indicate that copper deficiency triggers changes in the expression of genes encoding key enzymes in various chloroplast-localized lipid/pigment biosynthetic pathways. Among these are CGL78 involved in chlorophyll biosynthesis and HPPD1, encoding 4-hydroxyphenylpyruvate dioxygenase catalyzing the committed step of plastoquinone and tocopherol biosyntheses. Copper deficiency in wild-type cells does not change the chlorophyll content, but a survey of chlorophyll protein accumulation in this situation revealed increased accumulation of LHCSR3, which is blocked at the level of mRNA accumulation when either CGL78 expression is reduced or in the crd1 mutant, which has a copper-nutrition conditional defect at the same step in chlorophyll biosynthesis. Again, like copper-deficient crd1 strains, cgl78 knock-down lines also have reduced chlorophyll content concomitant with loss of PSI-LHCI super-complexes and reduced abundance of a chlorophyll binding subunit of PSI, PSAK, which connects LHCI to PSI. For HPPD1, increased mRNA results in increased abundance of the corresponding protein in copper-deficient cells concomitant with CRR1-dependent increased accumulation of γ-tocopherols, but not plastoquinone-9 nor total tocopherols. In crr1 mutants, where increased HPPD1 expression is blocked, plastochromanol-8, derived from plastoquinone-9 and purported to also have an antioxidant function, is found instead. Although not previously found in algae, this metabolite may occur only in stress conditions.
Keywords: antioxidant, Chlamydomonas, chlorophyll, light-harvesting complex (antenna complex), metal homeostasis, LCAA, SBP domain, Ycf54
Introduction
Copper is an essential cofactor for most forms of life because of its function as a catalyst of oxygen chemistry and redox reactions (1). In photosynthetic organs of land plants or in unicellular phototrophs like algae and cyanobacteria, plastocyanin is a major copper-requiring protein (2). Many algae and cyanobacteria reduce their dependence on copper (the copper quota) by replacing plastocyanin with a heme protein called cytochrome (Cyt)4 c6 (reviewed in Ref. 3). In Chlamydomonas, this switch is controlled by copper response elements associated with the CYC6 gene encoding Cyt c6 and a transcription factor, copper response regulator 1 (CRR1) (4–6). CRR1 has a SQUAMOSA promoter binding protein (SBP) domain that is conserved in the green lineage (6). Its homologue in Arabidopsis, SPL7, controls nutritional copper homeostasis as well (7, 8). Transcriptome experiments revealed multiple targets of this transcription factor in both organisms but besides the genes encoding assimilatory copper transporters (COPT/CTR), which are dramatically up-regulated in both organisms under copper deficiency (9, 10), the target genes are distinct (8, 11). This may be because modification of the photosynthetic apparatus is a key response in Chlamydomonas but not in Arabidopsis.
We noted previously that redox proteins, especially O2-dependent metabolic enzymes, are enriched among the CRR1 targets in Chlamydomonas (11). Among these are CRD1 and CPX1, encoding enzymes regulating two rate-limiting steps in tetrapyrrole biosynthesis: crd1 mutants are chlorotic in copper-deficient cells because of reduced chlorophyll (Chl) content accompanied by reduced abundance of PSI and LHCI (12, 13). The abundance of CAO1 mRNA encoding another O2-dependent enzyme in Chl biosynthesis is also increased in copper deficiency but this may occur independently of CRR1 (11). The motivation for regulation of tetrapyrrole biosynthesis is not clear, but one hypothesis is that copper deficiency necessitates modification of photosystem biogenesis pathways. FAB2, another CRR1 target, encodes an acyl-ACP desaturase, whose activity is required for the synthesis of unsaturated fatty acids in the chloroplast. Indeed, the galactolipids of the thylakoid membrane from copper-deficient cells are more unsaturated relative to those from copper-replete cells (11). Again, the underlying significance is not known, but because of the importance of unsaturated fatty acids for thylakoid membrane function, we hypothesized that replacement of plastocyanin with Cyt c6 may require structural modifications of the membrane.
In this work, we probe the function of two other CRR1 target genes encoding enzymes in pathways affecting pigment and lipid metabolism in the chloroplast, CGL78 (also called LCAA and Ycf54 in Arabidopsis/tobacco and cyanobacteria), a conserved protein that has been proposed to function with CRD1 in tetrapyrrole biosynthesis at the Mg Protoporphyrin IX monomethyl cyclase step (14–16), and HPPD1, encoding one of two isoforms of 4-hydroxyphenylpyruvate dioxygenase (HPPD), which catalyzes the conversion of 4-hydroxyphenylpyruvate to homogentisate (17). Although HPPD is critical for tyrosine catabolism in animals, in plants, this enzyme catalyzes the rate-limiting step of an anabolic branch that forms plastoquinone-9 (PQ-9) and tocopherols. PQ-9 is the mobile carrier of 2 reducing eq between PSII and the Cyt b6f complex, whereas tocopherols have antioxidant activity by scavenging singlet oxygen produced by excess excitation energy input into PSII and protecting membrane lipids from peroxidation (18–21). There are four tocopherol types, α-, β-, γ-, and δ-tocopherols, based on the number and position of methyl groups on the chromanol ring. The increase in abundance of α-tocopherols in response to high light is well documented for both algae and plants (22–24), and there is also a role for tocopherols in signaling pathways, although the mechanistic aspects are not yet fully elucidated (25).
For CGL78, we used an artificial micro-RNA-mediated reverse genetics approach to dissect its function, whereas for HPPD, we monitored pathway end products under conditions affecting its expression to validate its contribution to plastoquinone and tocopherol biosynthesis. We discovered: 1) that LHCSR3 accumulation is increased in copper-deficient cells and 2) that LHCSR3 accumulation was especially sensitive to loss of CGL78 as was the accumulation of PSAK, a connector between PSI and LHCI. This phenotype is similar to that of the crd1 mutant, which is blocked at the same step in Chl biosynthesis. For HPPD, we noted 3) that, an increase of HPPD1 is, surprisingly, not accompanied by an increase in all end products of the tocopherol/plastoquinone biosynthesis pathway. Rather, copper-deficient cells accumulate specifically γ-tocopherols. Moreover, when HPPD1 expression is blocked, as in the crr1 mutant, γ-tocopherol accumulation is abolished as well, supportive of a causal connection between HPPD abundance and γ-tocopherol accumulation. We speculate that operation of the light reactions is less effective in copper-deficient cells, requiring subtle modifications of mechanisms for handling excess excitation energy and stress.
Results
Reduced CGL78 Expression Impacts PSI/II-LHC Super-complex Formation
To assess the function of CGL78 in Chlamydomonas, we used an inducible amiRNA system to generate strains with conditionally reduced abundance of CGL78 mRNA. Because the amiRNA is driven by the NIT1 promoter, it is ammonium repressible (26). Therefore, strains cgl78-ami11 and cgl78-ami13 carrying the knock-down constructs accumulate CGL78 mRNA at normal abundance in medium with ammonium but have reduced abundance in medium lacking ammonium (8 and 5% as compared with control strains, CGL78, respectively (Fig. 1A). Accordingly, the cgl78-ami strains are chlorotic on ammonium-free medium (Fig. 1B) with significantly reduced Chl content relative to CGL78 (36%), although the ratio of Chl a to b is unaffected (Fig. 1C). There is no effect of cgl78-ami knock-down on growth (Fig. 1D), making it unlikely that loss of CGL78 affects any other pathway. The chlorotic phenotype indicated a direct role of CGL78 in chlorophyll-binding protein accumulation, but the photosystem II capacity was unaffected in cgl78-ami lines (Fig. 1E). To distinguish which chlorophyll-binding proteins might be reduced in cgl78-ami strains, we analyzed thylakoid membrane proteins after separation by BN-PAGE, which resolves various photosystem-containing complexes (27). We noticed a striking loss of PSI/II-LHC super-complexes (marked with a red arrow) with a corresponding increase in PSII core monomers (Fig. 2, compare unfilled with filled arrows).
FIGURE 1.
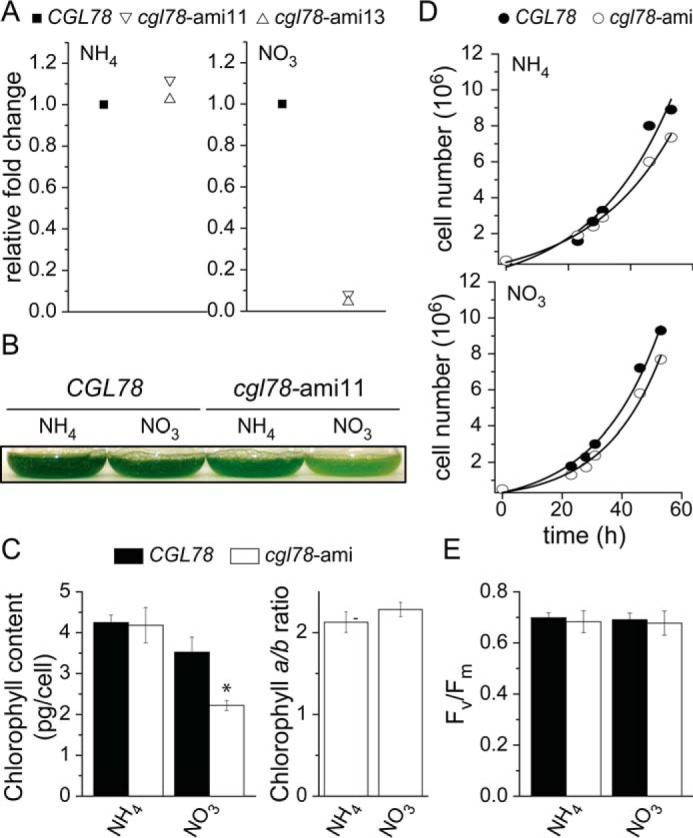
Loss of CGL78 expression results in a chlorotic phenotype. A, abundance of CGL78 mRNA of independent cgl78-amiRNA lines (11, 13) was determined by quantitative RT-PCR. RNA was isolated from cells that were either grown on TAP media containing ammonia or nitrate as the sole nitrogen source. B, representative picture of the pale green phenotype of cgl78-ami11 at a density of 8 × 106 cells/ml. C, chlorophyll content and chlorophyll a/b ratio. The averages and S.D. of different cgl78-ami lines are shown. D, representative growth curves of cgl78-ami11 and an empty vector control line (CGL78) that were grown in media containing either ammonia or nitrate as sole nitrogen source. Cells were grown in TAP medium containing ammonium (NH4+) to repress the amiRNA construct or nitrate (NO3) to induce the amiRNA construct. Experiments were done at least twice. E, the maximum PSII capacity was measured in independent CGL78 or cgl78-ami lines. Shown are the averages of three independent cultures ± S.D.
FIGURE 2.
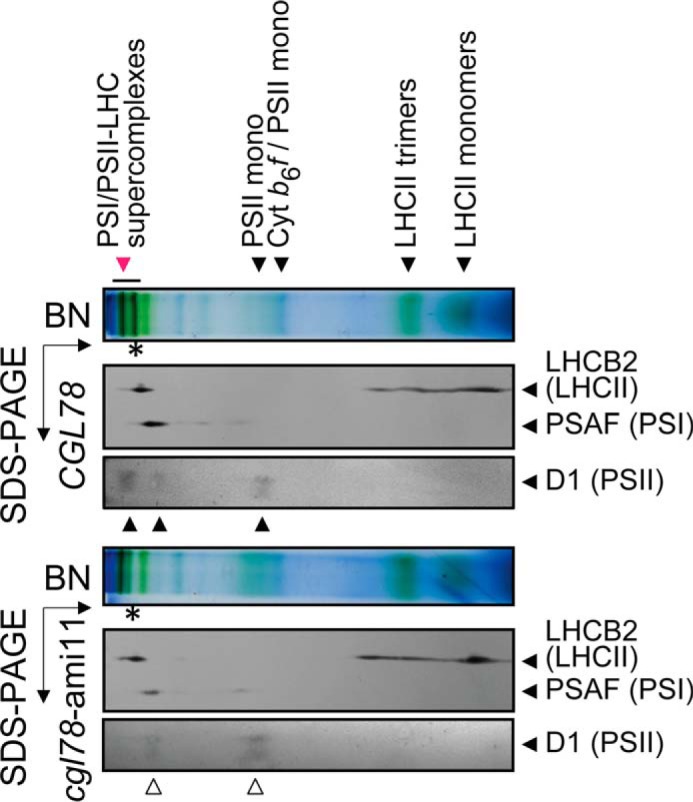
PSI/II-LHC super-complexes are reduced when CGL78 expression is down-regulated. Thylakoid membranes extracted from control (CGL78) or cgl78-ami lines reveal reduced chlorophyll content in PSI/II-LHC super complexes (red arrow). Super-complex composition of selected proteins was verified by second dimension, denaturing SDS-PAGE. Shown are immuno-detections with anti-LHCB2 (LHCII marker) and anti-PSAF (PSI marker) and anti-D1 (PSII marker). All experiments shown were performed at least twice either with cgl78-ami11 or cgl78-ami13.
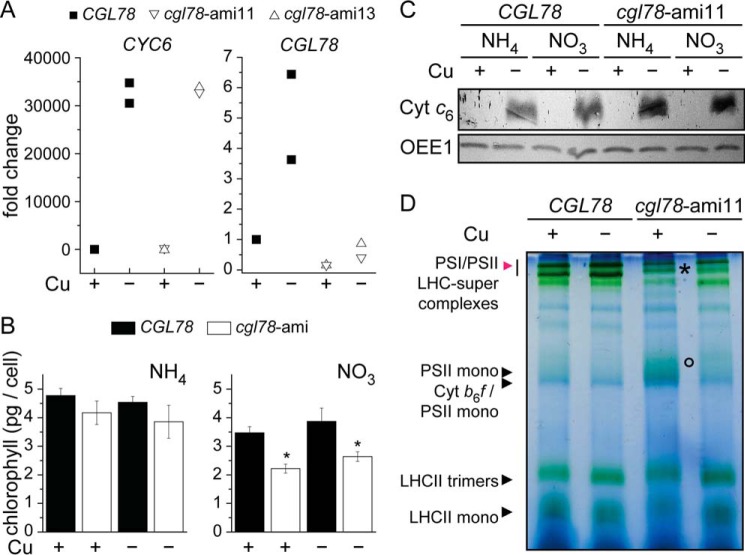
Because CGL78 is a target of CRR1 and both mRNA and protein abundances are increased 7- and 4-fold, respectively, in copper-deficient medium (Refs. 11 and 28 and Fig. 3A), we tested whether the cgl78-ami phenotype was affected by copper nutrition status. The copper status was verified by expression of the previously characterized sentinel gene CYC6 and accumulation of the corresponding protein (29) (Fig. 3, A and C). Nevertheless, copper neither exacerbated nor ameliorated the phenotype (Fig. 3B). Separation of chlorophyll-binding proteins by BN-PAGE revealed that copper deficiency did not affect the PSI/II-LHC super-complex formation in wild-type lines (Fig. 3D, red arrow), but copper-deficient cgl78-amiRNA lines showed reduced amounts of PSI/II-LHC super-complexes (Fig. 3D, marked by an asterisk). The pattern of PSI/II-LHC super-complexes of thylakoids isolated from cgl78-ami lines in copper-depleted versus copper-replete grown cells look similar except that PS core subunits are more enriched in the former (Fig. 3D, circle). On the other hand, accumulation of PSII was the same in cgl78-ami strains and control lines based on the abundance of D1 (Fig. 4).
FIGURE 3.
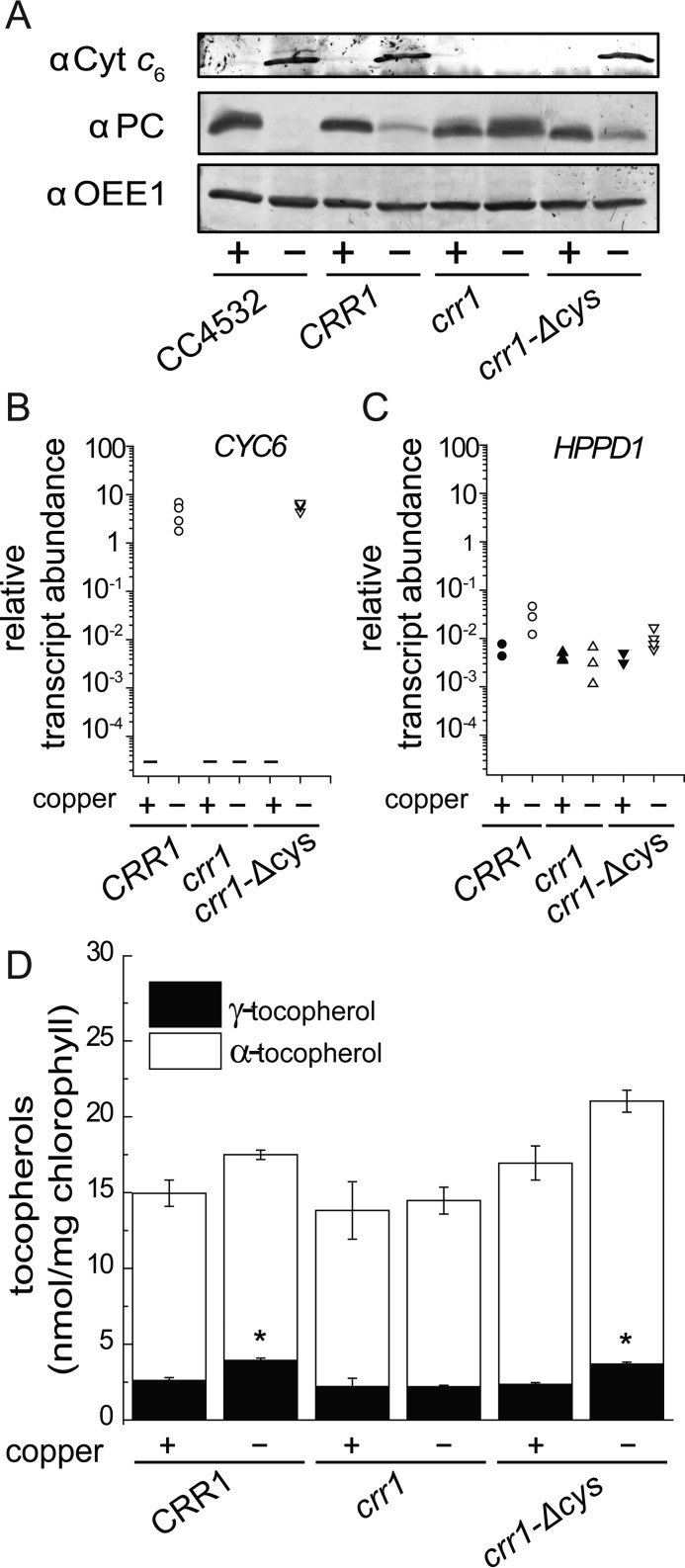
Copper deficiency does not impact the phenotype of cgl78-ami strains. A, CYC6 and CGL78 mRNA abundance was estimated by qRT-PCR. RNA was isolated from cells that were grown in medium containing nitrate to induce the amiRNA targeting CGL78, the presence of Cu(II)-EDTA in the medium is indicated. Control strains (black) or cgl78-ami lines (white) were grown in TAP medium to a density of 5–8 × 106 cells/ml. Each symbol represents an independent experiment analyzed in technical duplicates. B, chlorophyll content in cells grown in medium supplemented either with ammonia or nitrate as nitrogen source and supplemented with or without copper as indicted. Error bars represent S.D. of at least three experiments. C, soluble fractions corresponding to 10 μg of protein were separated on sodium dodecyl sulfate-containing denaturing gels (15% acrylamide) and immuno-detection with anti-Cyt c6 or OEE1 as a loading control. D, PSI/II-LHC super-complex formation is also diminished in copper-depleted cgl78-ami lines. Blue native PAGE of thylakoid membranes from CGL78 or cgl78-ami lines grown either in copper-replete or copper-depleted TAP medium with nitrate as nitrogen source. Experiments were performed at least twice.
FIGURE 4.
PSAK and LHCSR3 accumulation is dependent on CGL78. A, the abundance of the α and β subunits of CF1, appearing as a single band, were visualized as a loading control (bottom panel). Membrane fractions, equivalent to 10 μg of protein, were loaded on sodium dodecyl sulfate-containing denaturing gels. B, samples used for panel A were re-evaluated for estimating relative abundances. An asterisk marks the copper-replete sample, whereas the circle represents the copper-depleted sample from either cgl78-ami11 cells that were grown in medium containing nitrate as nitrogen source. C, shown are the relative mRNA abundances of CRD1, CTH1 (2.1 kb functional transcript), CTH1 (3.1 kb non-functional transcript) in either CGL78 (black squares) or cgl78-ami11 (white apex-down triangle or cgl78-ami13 (white apex-up triangle) that were grown in copper-depleted or copper-replete medium as indicated. D, CGL78 mRNA abundance in crd1 (white circles) or CRD1 (gray circles) was determined by qRT-PCR. Each symbol represents the average of a duplicate qPCR of an independent experiment.
In previous work, we observed that PSI-LHCI complexes are destabilized in iron-deficient cells, coincident with a loss of PSAK, which is the subunit connecting PSI and LHCI. The loss of PSI-LHCI complexes prompted us to monitor PSAK accumulation in cgl78-ami lines, where we found it to be decreased to 50% (Figs. 4 and 6C). This molecular phenotype is reminiscent of the crd1 mutant (30). CRD1 is one of two isoforms of a subunit of Mg Protoporphyrin IX monomethyl cyclase. Its expression is up-regulated by poor copper nutrition via CRR1 (12). The other isoform, named CTH1, is reciprocally regulated, also by CRR1 (13). Because CGL78 functions in concert with CRD1 and CTH1, we tested whether CRD1 and CTH1 expression might be affected in the cgl78-ami lines (Fig. 4C). We did not see any significant change in CRD1/CTH1 abundances in cgl78-ami lines (Fig. 4, A and B) nor in the pattern of mRNA accumulation as assessed by qRT-PCR (Fig. 4C). Likewise, CGL78 mRNA abundance is also not changed in crd1 as compared with CRD1 cells (Fig. 4D). Therefore, the expression of CGL78 seems not to be interconnected with that of CTH1/CRD1 and we can attribute the cgl78 molecular phenotype in copper deficiency directly to loss of CGL78 rather than indirectly to loss of its partner protein(s) in the complex.
FIGURE 6.
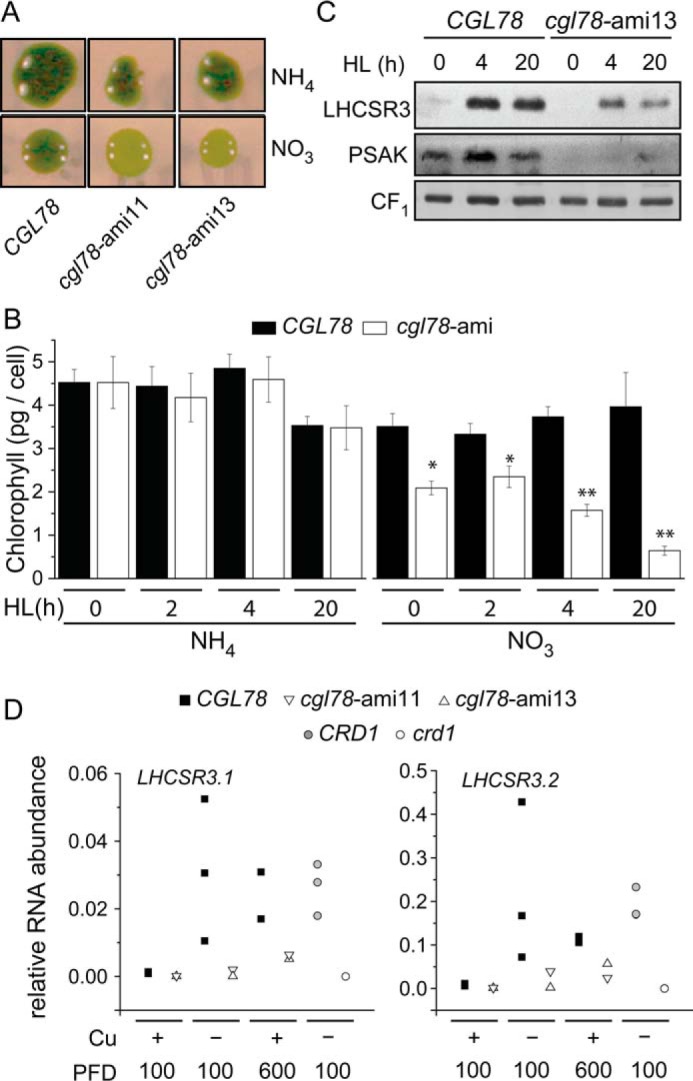
cgl78-ami strains are more sensitive to high light. A, colonies of CGL78 or cgl78-ami lines were either grown on TAP agar medium containing ammonia or nitrate as the sole nitrogen source for 1 week in low light (40 μmol m−2 s−1) and then exposed for 24 h to high light (600 μmol m−2 s−1). B, chlorophyll content of whole cells. Cells were either grown in TAP liquid medium containing nitrate (NO3) or ammonium (NH4) and transferred at a density of 2 × 106 cells/ml to high light (HL, 600 μmol m−2 s−1). Shown are the averages and S.E. of four independent experiments. Asterisks indicate a significant difference compared with CGL78 according to one-way analysis of variance, post hoc Holm-Sidak (*, p value < 0.05; **, p value < 0.001). C, protein was extracted from cells that were exposed to high light and grown in TAP medium containing nitrate. Membrane fractions, equivalent to 10 μg of protein, were loaded on sodium dodecyl sulfate-containing denaturing gels. The abundance of CF1 was used as a loading control (bottom panel). Shown is one of two independent experiments. D, LHCSR31 and LHCSR3.2 mRNA abundance in CGL78 (black squares), cgl78-ami11 (white apex down), cgl78-ami13 (white apex up), crd1 (white circles), or CRD1 (gray circles) was determined by qRT-PCR. Shown are the relative mRNA abundances of CRD1, CTH1 (2.1 kb fragment), and CTH1 (3.1 kb fragment). Cells were grown in copper-replete (+) or copper-depleted (−−) medium in continuous light at a PFD of 100 μmol m−2 s−1 or exposed to 24 h high light (600 μmol m−2 s−1) as indicated. Each symbol represents the average of a duplicate qRT-PCR of an independent experiment.
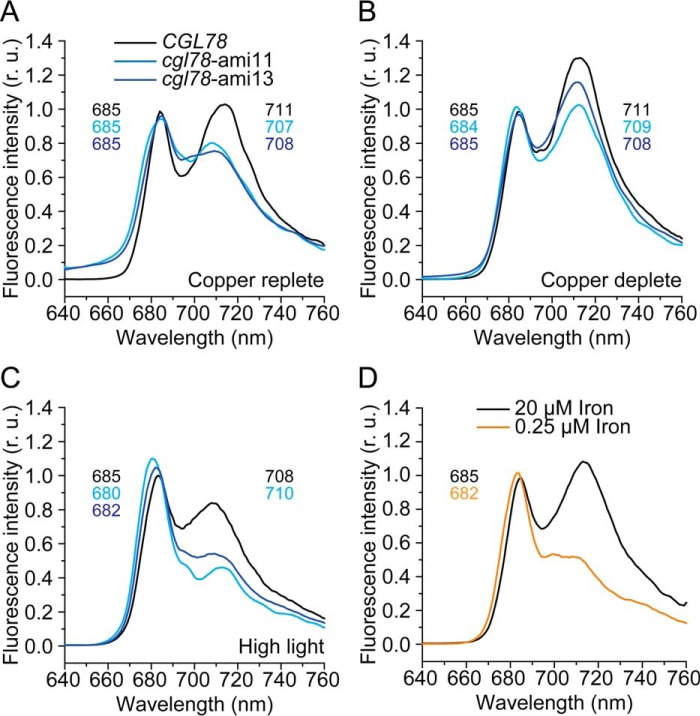
In previous work, we showed that the presence of PSAK was related to functional association of the LHCI antenna with PSI (30). For instance, in iron-deficient cells, which are impaired in chlorophyll biosynthesis, PSAK is reduced, resulting in less energy transfer from LHCI to PSI. Therefore, we collected fluorescence emission spectra to assess PSI-LHCI interactions in wild-type versus knock-down lines as a function of copper nutrition. The copper-replete control cells (CGL78) showed characteristic peaks at 685 and 711 nm, resulting from fluorescence of LHCII attached to PSII and of LHCI to PSI, respectively (Ref. 31 and Fig. 7A). Copper-depleted control cells showed the same changes of the emission spectra that we had observed in copper-depleted wild-type cells in previous work (13), namely an increase in intensity of the peak at 710 nm relative to the peak at 685 nm, indicating modifications of the PSI-LHCI interaction in copper-deficient cells (Fig. 7B). In cgl78-ami lines, the PSI-LHCI peak at 710 nm was blue-shifted (32) and reduced in intensity under both, copper-depleted and copper-replete conditions (Fig. 7, A and B). The latter is consistent with the observation that PSAK is reduced in cgl78-ami lines. When wild-type strains are grown in high light, we note reduced intensity and a blue-shift of the peak corresponding to PSI-LHCI (Fig. 7C), whereas cgl78-ami lines subject to high light show a blue-shift of the PSII-LHCII peak to 682 nm and loss of the PSI-LHCI peak (Fig. 7C). The latter situation is reminiscent of iron-starved cells (Ref. 30 and Fig. 7D), and is consistent with a complete loss in both cases of PSAK (Ref. 30 and Fig. 6C).
FIGURE 7.
Weaker antenna-reaction center association in copper-deficient cells. 77 K fluorescence emission spectra of samples prepared from CGL78 in black, cgl78-ami11 in bright blue, and cgl78-ami13 in dark blue. Cells are grown in TAP medium with copper (+Cu) (A) or without copper (−Cu) supplementation (B) at a PFD of 100 μmol m−2 s−1. Cells grown in TAP medium with copper were exposed for 24 h to high light (600 μmol m−2 s−1) (C). D, CGL78 lines were grown in TAP medium containing 20 (black) or 0.25 μm (orange) iron. The excitation wavelength was 435 nm. Fluorescence emission was normalized to the value at 685 nm. The peaks are labeled with the wavelength of the maximum if applicable.
Copper-deficient Cells Accumulate a Stress-related Antenna Protein, LHCSR3
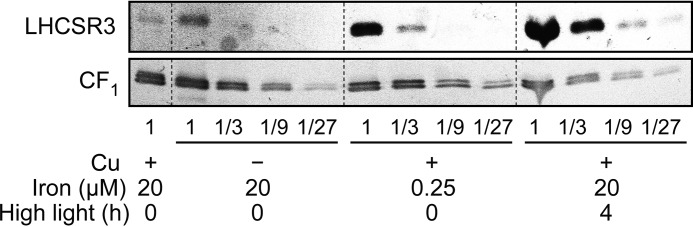
Besides the change in PSI-LHCI interactions noted above, a survey of chlorophyll-protein accumulation revealed a previously unrecognized difference between copper-depleted and copper-replete cells. Specifically, LHCSR3 is increased in copper-deficient wild-type (CGL78) cells severalfold, but not in copper-deficient cgl78-ami strains (Fig. 4). Quantification by a dilution series indicates that LHCSR3 abundance in cgl78-ami strains is about 12.5% of the level in the control strains (Fig. 4, A and B).
When we surveyed the copper-deficiency transcriptome relative to the iron-deficiency transcriptome, where up-regulation of LHCSR3 has already been described (33), we noted a small but consistent increase in LHCSR3.1 and LHCSR3.2 mRNA abundance in copper-deficient relative to copper-replete cells (Table 1). The change is smaller than that documented for iron-starved cells (33) and substantially less than that in cells exposed to high light, but it is statistically significant. When we compared the abundance of LHCSR3 in copper-deficient cells to the abundance in iron-limited cells, again the increase was correspondingly lower in the copper-deficient situation (Fig. 5). A simple explanation might be that copper-deficient cells are also secondarily iron-deficient, but measurements of the iron content in copper-deficient cells indicate that the iron content in copper-replete cells (10.6 ± 1.4 × 107 atoms/cell) does not differ from the iron content in copper-depleted cells (10.4 ± 1.1 × 107 atoms/cell). Therefore, we conclude a direct consequence of copper nutrition on the performance of the PSI-LHCI complex.
TABLE 1.
LHCSR3 expression is up-regulated in copper-depleted and iron-limiting conditions
Shown is the expression in fpkm (fragments per kilobase of exon per million fragments mapped) of LHCSR3.1 (Cre08.g367500), LHCSR3.2 (Cre08.g367400), and the total amount of LHCSR3-encoding mRNA in the condition indicated. For the light experiments cells were grown for 12 h in the dark and then shifted for 4 h to light. Data were obtained from Refs. 11 and 33.
| Gene | Condition |
|||||
|---|---|---|---|---|---|---|
| Copper |
Iron |
Light |
||||
| 2 | 0 | 20 | 0.25 | 0 | 250 | |
| μm | μm | μmol m−2 s−1 | ||||
| LHCSR3.1 | 0.4 | 2.3 | 0.1 | 1 | 0.3 | 48 |
| LHCSR3.2 | 1.4 | 3.3 | 0.3 | 3.2 | 0.2 | 27.4 |
| Total | 1.8 | 5.6 | 0.4 | 4.2 | 0.5 | 75.4 |
FIGURE 5.
LHCSR3 increase in copper-deficient cells compared with iron limited and high light-exposed cells. Change in abundance of LHCSR3 in response to growth in copper-deficient medium, iron-deficient medium or high light conditions. The abundance of CF1 was visualized as a loading control (bottom panel). Membrane fractions, equivalent to 10 μg of protein, were loaded on sodium dodecyl sulfate-containing denaturing gels.
Because LHCSR3 seemed to be the most affected chlorophyll-binding protein in copper-deficient cgl78-ami lines, and LHCSR3 has a known role in photo-protection (34–38) we wondered if cgl78-ami strains are more sensitive to high light relative to the control. Indeed, if we grew cgl78-ami lines in high light, we observed that the cells became bleached (Fig. 6A). The chlorophyll content of cgl78-ami strains that were grown for 20 h in high light is reduced to 5.5%, as compared with CGL78 wild-type lines (Fig. 6B), concomitant with a strong reduction of LHCSR3 abundance and nearly complete absence of PSAK (Figs. 6C and 7, A and D). Reduced abundance of LHCSR3 likely occurs at the level of mRNA accumulation because the increased LHCSR3 expression in copper deficiency is blocked in the cgl78-ami lines. In fact, the high light responsive increase in LHCSR3 mRNA is also blocked in the cgl78-ami lines (Fig. 6C). This prompted us to test LHCSR3 expression in the crd1 mutant, and we noted that this molecular phenotype is recapitulated in crd1. The latter suggests that the cyclase in Chl biosynthesis might signal to the LHCSR3 gene. Another possibility is that a functional electron transport chain in PSI is required for increased LHCSR3 expression (39).
Tocopherol Metabolism Is Altered in Copper-deficient Chlamydomonas
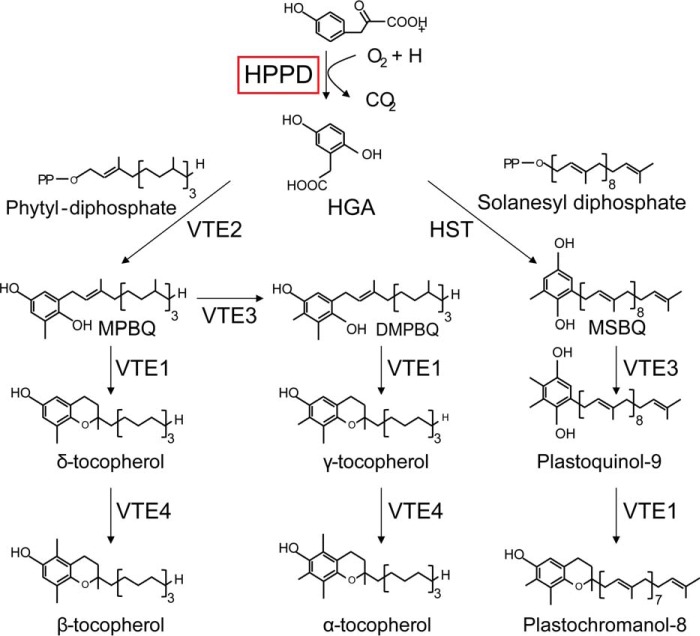
Besides Chl biosynthetic enzymes, previous transcriptome analyses had identified other changes in cofactor metabolizing enzymes in copper-deficient cells. One of these is HPPD1, a likely target of CRR1, and one of two genes encoding HPPD, which catalyzes the first step in a branched pathway leading to the terpenoid cofactors, plastoquinone and tocopherols (Fig. 8). Sequence analysis indicates that both genes are more highly related to homologues in plants versus animals. The two isoforms are paralogues; they are most closely related to algal homologues but even more closely related to each other (90% identity at the nucleotide level and 93.5% similarity at the protein level) consistent with a recent duplication event. Previous transcriptome and proteome profiling indicated that HPPD1 mRNA and the corresponding protein are increased in copper-deficient cells and in cells that experienced hypoxia (11, 28) (Fig. 9A), which was validated in this work by real time PCR with paralog-specific primers and immunoblotting (Fig. 9, B and C). Antibodies raised against the carrot protein recognized an approximate 46-kDa protein whose abundance is increased in copper-deficient relative to copper-replete Chlamydomonas cells (Fig. 9C). The impact of CRR1-dependent regulation is an ∼2-fold increase in mRNA templates encoding HPPD in copper-deficient growth conditions (Fig. 10B). Given the function of these molecules in electron transfer and photoprotection, we measured the abundance of the end products of the pathway to assess the impact of increased expression.
FIGURE 8.
Overview of the plastoquinone and tocopherol biosynthesis pathway in Chlamydomonas. HGA, homogentisate; HST, homogentisate solanesyl transferase; DMPBQ, 2,3-dimethyl-5-phytyl-1,4-benzoquinone; HPPD, 4-hydroxyphenylpyruvate dioxygenase; MPBQ, 2-methyl-6-phytyl-1,4-benzoquinol; MSBQ, 2-methyl-6-solanesyl-1,4-benzoquinol; VTE1, tocopherol cyclase; VTE2, homogentisate phytyltransferase (HPT); VTE3, 2-methyl-6-phytylplastoquinol methyltransferase (MPBQMT); VTE4, γ-tocopherol methyltransferase (γ-TMT).
FIGURE 9.
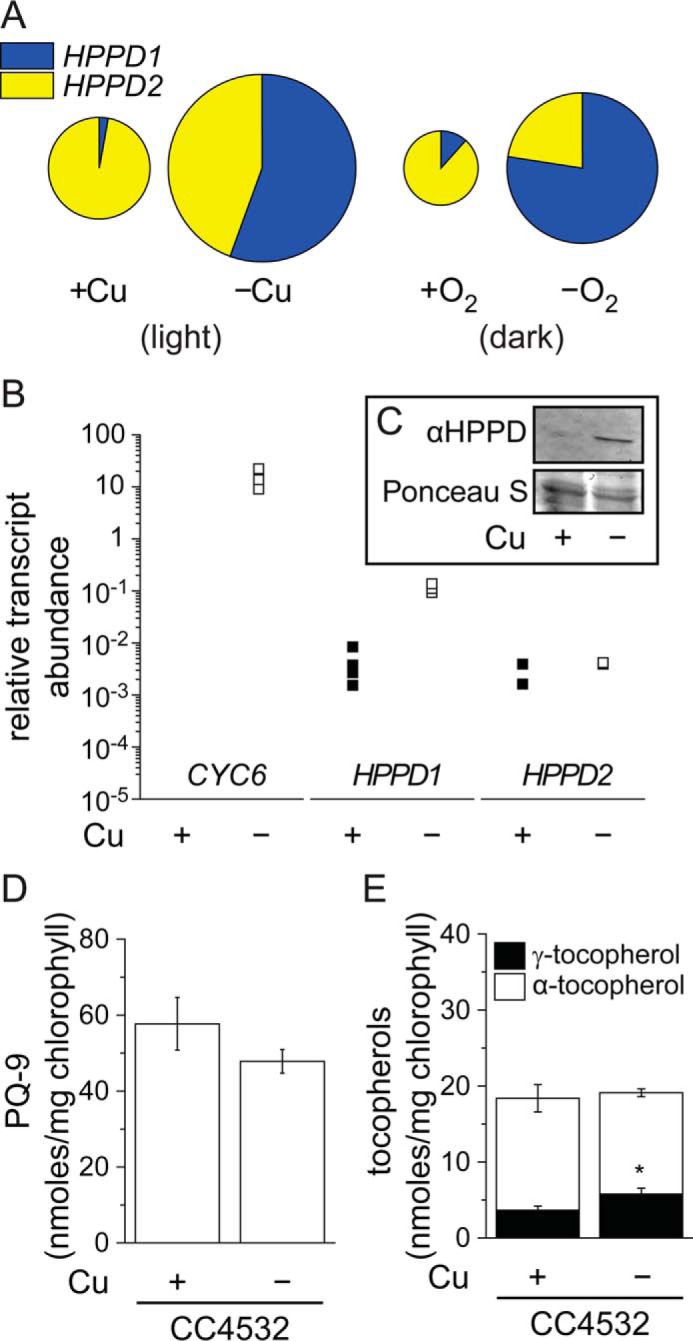
HPPD accumulation correlates with HPPD1 abundance and increased γ-tocopherol in copper-deficient Chlamydomonas. A–D, change in HPPD-encoding transcripts in response to growth in copper-deficient medium or dark anoxic conditions. A, data from RNA-seq experiments in Chlamydomonas strain CC4532. The sizes of the circles are proportional to the relative abundance (RPKM) of the total HPPD transcript pool in each condition. B, abundance of CYC6, HPPD1, and HPPD2 transcripts was also estimated in independent samples by qRT-PCR. CC4532 was grown in TAP medium in the condition indicated: −Cu(II) (black squares) or +Cu(II) (white squares). Cells were collected after reaching a density of 5–8 × 106 cells/ml and analyzed for RNA abundance by real-time PCR. Each symbol represents an independent experiment analyzed in technical duplicates. C, proteins were extracted from copper-depleted or copper-replet conditions and 10 μg of soluble protein was separated by 10% PAGE, followed by immuno-detection with an antibody against Arabidopsis HPPD. D, plastoquinone, and E, α- and γ-tocopherol contents were measured in extracts of Chlamydomonas cells. Data are the averages of three experimental replicates ± S.D.
FIGURE 10.
The increase of γ-tocopherol content is dependent on CRR1. A, for immuno-detection of plastocyanin and cytochrome c6, soluble proteins were separated by SDS-polyacrylamide gel electrophoresis (15%) and transferred to a nitrocellulose membrane (0.1 n pore size). The abundance of OEE1 was used as a loading control (bottom panel). B and C, abundance of CYC6 (B) and HPPD1 (C) transcripts. Independent cultures corresponding to CRR1 (black circles), crr1 (triangles apex up), or crr1-ΔCys (triangles apex down) were grown in copper-replete (black symbols) or copper-depleted (white symbols) TAP medium. Cells were collected after reaching a density of 2–6 × 106 cells/ml and analyzed for RNA abundance. Each symbol represents an independent experiment analyzed in technical triplicates. D, the tocopherol content in Chlamydomonas crr1 and corresponding strains that have been complemented with full-length CRR1 (crr1(CRR1)) or with a truncated version of CRR1 lacking a cysteine-rich domain (crr1-ΔCys) was measured as described under “Experimental Procedures.” Data are the averages of three biological replicates ± S.D.
Surprisingly, despite the increase in mRNA and protein at a rate-limiting step, the total plastoquinone or tocopherol content was not changed in copper-deficient versus copper-replete Chlamydomonas (Fig. 9, D and E). Nevertheless, we noted a consistently higher proportion of γ-tocopherol (42% increase on average) in copper-deficient cells of strain 4532 (wild-type for CRR1) (Fig. 9E). To evaluate the contribution of HPPD1 to γ-tocopherol content, we took advantage of the crr1 mutant (because HPPD1 is a CRR1 target) and two independent complemented lines, CRR1 (in which the wild-type gene rescued crr1) and crr1-ΔCys (in which a gene encoding a C terminally truncated version of CRR1 complements the crr1 mutation). The copper status of the cultures was validated on the basis of sentinel protein accumulation (Fig. 10A). Both complemented lines, but not the crr1 mutant, show normal CRR1- and copper-responsive expression of HPPD1 and the sentinel CYC6 gene (Fig. 10, A–C). The increase in γ-tocopherol mirrors CRR1 function, namely observed in both complemented lines but not in the crr1 mutant (Fig. 10D). Therefore, we conclude that the change in the γ-tocopherol to α-tocopherol ratio is dependent on copper nutrition and CRR1 and is an intrinsic response to poor copper nutrition.
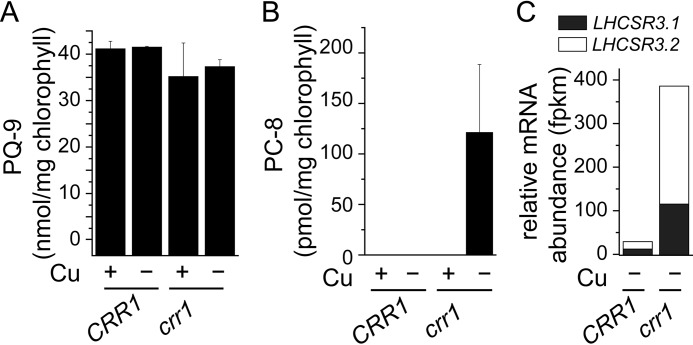
When we surveyed plastoquinone species in crr1 versus CRR1, we found that the total PQ-9 content was unaffected (Fig. 11A), consistent with a lack of effect of copper nutrition on plastoquinone content, but we did observe a new metabolite, plastochromanol-8 (PC-8), previously not observed in algae (Fig. 11B). PC-8 is a stress metabolite generated from plastoquinol-9 by a cyclization reaction catalyzed by VTE1. We hypothesize that PC-8 accumulation in copper-deficient crr1 is part of a general stress acclimation response in this situation because it is independent of an increase in HPPD expression. Its abundance relative to PQ-9 is small (0.35% of total) and is found only in copper-deficient crr1 cells, a situation in which HPPD1 is not up-regulated and γ-tocopherol increase is not observed. One possibility is that PC-8 is produced for a photo/ROS-protective function in copper-deficient crr1 strains, a hypothesis that is consistent with the up-regulation of LHCSR3 and reduced PSAK in this situation (Fig. 11C).
FIGURE 11.
Copper-deficient crr1 accumulates PC-8. A, plastoquinone or, B, plastochromanol content in Chlamydomonas crr1 and a strain that has been complemented with full-length CRR1 (crr1(CRR1)). Respective strains were either grown in medium with (+) or without copper (−) as indicated. Data are the averages of three biological replicates ± S.D. C, LHCSR3.1 (black) and LHCSR3.2 (white) transcript abundance was determined by RNA-seq in a copper-depleted crr1 and CRR1 (crr1(CRR1)) (11).
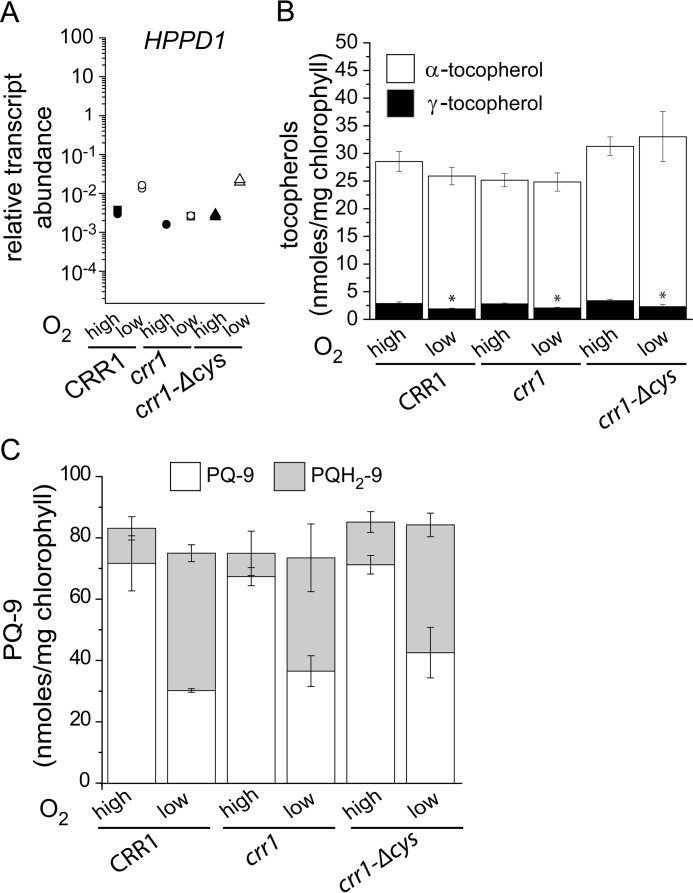
HPPD1 expression can be increased also in hypoxia in a CRR1-dependent pathway (40) (Fig. 12, A and B). Nevertheless, in this situation the enzymatic activity should actually be compromised because of low O2, a substrate of the reaction. Indeed, wild-type hypoxic cells have less γ-tocopherol than do normoxic cells (Fig. 12C, 66% reduced), which confirms that an oxygen-dependent step is required for the change in tocopherol composition beyond simply increased mRNA accumulation. When we tested crr1 mutants versus complemented strains, again the hypoxic increase in HPPD1 mRNA abundance is dependent on CRR1 (Fig. 13A), but in the absence of oxygen, γ-tocopherol content is decreased rather than increased relative to normoxic cells (Fig. 13B). When we looked at the plastoquinone profile in the hypoxic cells, we noted that the total PQ-9 content is unchanged in crr1 mutants or complemented strains (Fig. 13C), but the ratio of oxidized to reduced is dramatically reduced in hypoxic cells of all genotypes, which validates the physiology of the treatment.
FIGURE 12.
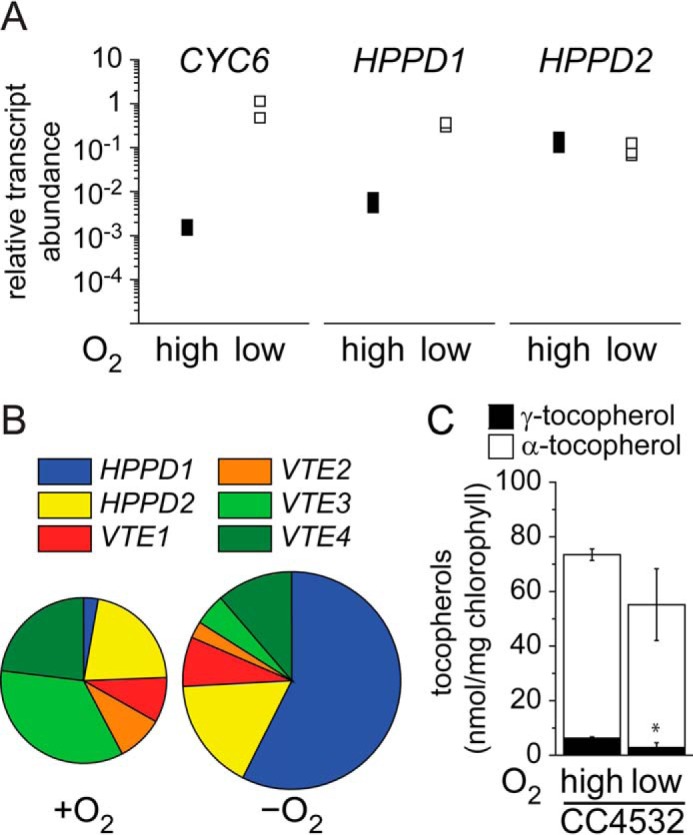
HPPD1 is also regulated by dark hypoxia but γ-tocopherol content is decreased in dark hypoxic Chlamydomonas. A, abundance of CYC6, HPPD1, and HPPD2 transcripts. Independent cultures corresponding to Chlamydomonas wild-type strain CC4532 were grown in dark hypoxia. Cells were collected after reaching a density of 2–6 × 106 cells/ml and analyzed for RNA abundance. Each symbol represents an independent experiment analyzed in technical triplicates. B, transcript abundance of genes encoding enzymes of the tocopherol biosynthesis pathway were determined by RNA sequencing in dark grown and dark anoxic Chlamydomonas strain CC4532 (38). The sizes of the circles are proportional to the relative abundance (RPKM) of the total transcript pool of genes encoding enzymes regulating the plastoquinone/tocopherol pathway as shown in Fig. 8. C, the tocopherol content in Chlamydomonas CC4532 that was either grown in the dark, normoxic (high) or in the dark, hypoxic (low) as indicated.
FIGURE 13.
The decrease of γ-tocopherol in dark hypoxia is independent of CRR1. A, abundance of HPPD1 transcripts as determined by quantitative real time PCR. Independent cultures corresponding to CRR1 (black circles), crr1 (triangles apex up), and crr1-ΔCys (triangles apex down) were grown in TAP medium in the conditions indicated: dark hypoxia (white symbols) relative to dark (black symbols). Cells were collected after reaching a density of 2–6 × 106 cells/ml and analyzed for RNA abundance. Each symbol represents an independent experiment analyzed in technical triplicates. Tocopherol (B) and plastoquinone (C) content in Chlamydomonas crr1 and corresponding strains that have been complemented with full-length CRR1 (crr1(CRR1)) or with a truncated version of CRR1 lacking a cysteine-rich domain (crr1-ΔCys). Respective strains were either grown in the dark, normoxic (high) or hypoxic (low) as indicated. Data are the averages of three biological replicates ± S.D.
Discussion
Acclimation to Copper Deficiency
Cyt c6 and Acyl-ACP Desaturase
Plastocyanin is the most abundant copper protein in photosynthetic cells and accordingly photosynthesis is dependent on copper in most organisms, except where there is a genetically programmed pathway to accommodate copper deficiency (41). The copper deficiency response occurs in many cyanobacteria and algae (42). The most well recognized change in the pathway of the light reactions in copper-deficient cells is the replacement of plastocyanin with a copper-independent substitute, a soluble c-type cytochrome or Cyt c6 (43–45). In this and previous work, we show that there are, in addition, subtler changes of the photosynthetic apparatus. These changes may be required to accommodate the use of structurally distinct mobile electron donors to PSI or to accommodate Cyt c6 as a less effective catalyst. Although it is not yet evident in the laboratory setting, the absence of Cyt c6 in land plants, which makes photosynthesis strictly copper dependent, suggests that plastocyanin offers a selective advantage.
Whole transcriptome analyses of copper-replete versus copper-deficient and CRR1 versus crr1 strains revealed dozens of genes with candidate copper-response elements that are likely targets of nutritional copper signaling (11). Many of these encode key enzymes in plastid lipid or lipid cofactor metabolism, such as coprogen oxidase (CPX1), CRD1 and CGL78 in tetrapyrrole biosynthesis, acyl-ACP desaturase (FAB2), and HPPD (discussed below). There are two possible rationales for increasing the expression of metabolic enzymes in copper deficiency: 1) increased expression is a compensatory mechanism that allows maintenance of end products because the pathway is compromised in copper-deficient cells or 2) increased expression allows increased accumulation of end products, more of which are required in copper-deficient cells. The two models are distinguished by end product analyses in wild-type versus crr1 mutants, and previous analyses showed that the latter is the situation for FAB2 and CRD1. Indeed, there are more unsaturated fatty acids in copper-deficient Chlamydomonas cells, whereas in the crr1 mutant the level of desaturation is unchanged (11). Lipid-profiling indicated that the enrichment was restricted to the galactolipids in the thylakoid membrane, one of the first indications that acclimation to copper deficiency requires modification of the photosynthetic membranes.
γ-Tocopherol and Plastochromanol-8
In this work, we document other subtle but measurable modifications to the photosynthetic apparatus. Increased expression of HPPD1 is (like for FAB2) causally connected with increased γ-tocopherol content. The proportion of γ-tocopherol is small (17% of the total tocopherol pool) and the increase therefore is also small, but it is reproducible and statistically significant, comparable with the magnitude of the change in the galactolipid desaturation. If Cyt c6 is a less effective catalyst relative to plastocyanin, the change in proportion of γ-tocopherol may serve to fine tune photosynthetic physiology by adjusting antioxidant or stress capacity. We conclude that like the situation for FAB2 and CRD1, increased expression of HPPD is required for increased accumulation of an end product (in this case γ-tocopherol).
Plastochromanol-8 has not previously been found in algae, but was revealed here in the crr1 mutant and only in copper deficiency where crr1 grows poorly. Because it is implicated in stress protection (46), its production may be a downstream consequence of stress in crr1 mutants. It would be interesting to test whether other stress situations also allow PC-8 accumulation and to distinguish the relevant regulatory mechanisms. In Arabidopsis, PC-8 levels are regulated by the activity of a type II NADPH dehydrogenase C1 (ortholog is NDA5 in Chlamydomonas) (47, 48) and a plastid ABC1-like kinase (49).
We had expected increased HPPD1 expression to yield increased total tocopherol, but this is not the case. It is possible that the biosynthetic pathways for α- versus γ-tocopherol in Chlamydomonas are branched at the level of suborganellar organization of enzyme complexes rather than at the level of the VTE3 enzyme. Increased expression of HPPD1 is not by itself sufficient to change the proportion of γ-tocopherol. In hypoxic cells, expression of HPPD1 is also increased but in the absence of the substrate O2, γ-tocopherol is not increased (Fig. 12C).
Chlorophyll-binding Proteins
We describe two other examples of adjustment of stress protection mechanisms in Chlamydomonas. First, copper-deficient cells have increased LHCSR3 content (Fig. 4). Although the increase is not as dramatic as in iron-deficient Chlamydomonas cells (50), it is notable, and more importantly, it is blocked in cgl78-ami lines and in crd1 mutants (Fig. 6C), suggestive of a signaling function of the cyclase with respect to expression of particular chlorophyll proteins. The operation of the tetrapyrrole pathway has been linked to nuclear gene expression in many works; this work documents a new regulatory connection (51).
LHCSR3 functions in photoprotection (35) involving photosystem I in Physcomitrella patens (52); its adjustment in copper-deficient cells may serve as pre-emptive fine tuning to accommodate Cyt c6 as a less effective donor to PSI relative to plastocyanin. The up-regulation of the tetrapyrrole pathway, documented for coprogen oxidase and the aerobic oxidative cyclase (CPX1, CRD1, CGL78) at the level of RNA and protein (28, 29) may allow adjustment of the abundance of specific chlorophyll-binding proteins to optimize the operation of the light reactions in a copper-deficient cell.
Second is de-stabilization of the LHCI-PSI interaction. Survey of photosystem I in the copper-replete situation showed a specific loss of PSAK, leading to de-stabilization of PSI-LHCI super-complexes. The cgl78-ami strains are also deficient in PSAK (Fig. 4), perhaps contributing to the phenotype noted by blue native gel separation of Chl-protein complexes and the observed blue-shift of the PSI-LHCI emission at 77 K. The loss of PSAK is even more dramatic in cgl78-ami lines subject to a high light treatment (Fig. 6). We had suggested previously that PSAK may modulate the connection between LHCI and PSI, consistent with its location in the complex (53), and hence serve a role in photo-protection.
We conclude that replacement of plastocyanin with Cyt c6 in response to copper deficiency requires considerable adjustments of the photosynthetic apparatus and photoprotective mechanisms. Although CGL78 is 5-fold up-regulated in copper-deficient cells, the cgl78-ami mutants are chlorotic independent of copper nutrition (Fig. 3). This contrasts with the copper nutrition-conditional crd1 phenotype, but this might be because CTH1, the reciprocally expressed paralog of CRD1, covers the loss of function in copper-replete situations.
Experimental Procedures
Strains and Culture Conditions
A miRNA targeting Chlamydomonas CGL78 was designed according to Refs. 26 and 54 using the WMD3 tool at wmd3.weigelworld.org. Resulting oligonucleotides CGL78amiFor, ctagtTAGATGTGGATCACGTACATAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTATGAACGTGATCCACATCTAg and CGL78amiRev, ctagcTAGATGTGGATCACGTTCATAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTATGTACGTGATCCACATCTAa (uppercase letters representing miRNA*/miRNA sequences) were annealed by boiling and slowly cooling down in a thermocycler and ligated into SpeI-digested pMS539, yielding pDS1. pDS1 was linearized by digestion with HindIII and transformed into Chlamydomonas strain CC4351 by vortexing with glass beads (55).
CGL78-amiRNA strains and Chlamydomonas reinhardtii strains CC4532, crr1–2 (CC5068), CRR1 (CC5071), and crr-Δcys (CC5073) (56) were grown in Tris acetate/phosphate (TAP) with constant agitation in an Innova incubator (180 rpm, New Brunswick Scientific, Edison, NJ) at 24 °C in continuous light (90 μmol m−2 s−1), provided by cool white fluorescent bulbs (4100 K) and warm white fluorescent bulbs (3000 K) in the ratio of 2:1, unless stated otherwise. High light (600 μmol m−2 s−1) was provided by a white fluorescent bulb (3800 K). TAP medium with or without copper was used with revised trace elements (Special K) instead of Hutner's trace elements according to Ref. 57. CGL78-amiRNA strains were either inoculated from a plate into standard TAP(NH4) to repress the artificial micro-RNA or a modified TAP medium, where nitrate was substituted instead of ammonium as the sole nitrogen source, TAP(NO3). Dark hypoxic cells were grown in TAP medium for 24 h in 1% air, 2% CO2, and 97% N2 by bubbling before collection as described in Ref. 56. Cell density (number of cells per milliliter of culture) was determined with a hemocytometer.
Chlorophyll Measurements
Chlorophyll was extracted from whole cells using an 80/20 (v/v) acetone/methanol mixture. Chlorophyll content was estimated according to Ref. 58 from the absorbances at 646.6 and 663.6 nm measured on a PerkinElmer LAMBDA 25 UV/visual spectrometer. The absorption at 750 nm was used as a reference.
RNA Extraction and cDNA Synthesis
2–4 × 107 cells were collected by centrifugation for 5 min at 2450 × g, 4 °C. RNA was extracted using the TRIzol reagent as described previously (59), treated with Turbo DNase (Ambion), concentrated, and cleaned with the Zymo Research RNA Clean & ConcentratorTM-5 Kit according to the manufacturer's instructions. Reverse transcription was primed with oligo(dT)18 using 2.5 μg of total RNA and SuperScript III (Invitrogen) according to the manufacturer's instructions.
Quantitative Real-time PCR
cDNA was diluted 10-fold before use. qRT-PCR contained 5 μl of cDNA, 6 pmol of each forward and reverse primer, 2 μl of Taq polymerase, 0.5 μl of 10 mm deoxynucleotide triphosphate (New England Biolabs), 2 μl of ×10 Ex Taq buffer (Mg2+ plus) (TaKaRa), 2 μl of ×10 SYBR mix (0.1% (w/v) SYBR Green 1 Nucleic Acid Gel Stain from Cambrex, 1% (w/v) Tween 20, 1 mg ml−1 of BSA, and 50% (v/v) DMSO) in a 20-μl volume. The following program was used: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 65 °C for 60 s. Fluorescence was measured after each cycle at 65 °C. A melting curve analysis was performed afterward from 65 to 95 °C with fluorescence reads every 0.5 °C. Relative abundances were calculated using LinReg. The abundance (No) of RACK1 (CBLP, Cre13.g599400) served as a reference transcript. Primers used in this study are as follows: CYC6For, CAGGTCTTCAACGGCAACTGT; CYC6Rev, ATCGCCCCCTTGCCAT, HPPD1For, GGTCGCGTCGATTGGGTTAC; HPPD1Rev, TGAGAACTCGTGGAAGCCACA; HPPD2For, ACCTCCTTCGGCCTGCAAC; HPPD2Rev, CACGTCCTCCGCAACAAACT; CGL78For, CCTGGACCGCGTGCTGAAGA; CGL78Rev,TACCGGGCGTAAGGGGCAGT; LHCSR3f, CACAACACCTTGATGCGAGATG; LHCSR3r, CCGTGTCTTGTCAGTCCCTG; CRD1For, CGTAGGTAGGCTGACTGCGTTG; CRD1Rev, GTCATTTATGCGCAGCCCTTG; CTH1(2)F, AGCTGTGCGTCGCGGGCTTGT; CTH1(2)R, ATCCGCGTGTTCCGAAGAAAC; CTH1(3)F, ACGCAGCAGCACAGCTCACT; and CTH1(3)R, TCCCCAAGTCTAGCCCGATG.
Thylakoid Membrane Preparation and Blue Native Gel Electrophoresis
Thylakoid membranes were prepared as described in Ref. 27 with the following modification: cells were collected by centrifugation at 3100 × g at 4 °C and re-suspended in 20 mm Na-Tricine, pH 8, 10 mm MgCl2, 0.4 m sorbitol; 0.2% BSA before sonication (two cycles of 5 s, 50% power). Thylakoid membranes were mildly solubilized for 15 min on ice in the dark with 1.5% β-dodecyl-D-maltoside (β-DM). The unsolubilized membranes were removed by centrifugation at 12,100 × g for 5 min, and the solubilized thylakoid membranes equivalent to 20 μg of chlorophyll were separated by BN-PAGE. For the second dimension, gel slices were incubated in 0.1% 2-mercaptoethanol, 10% glycerol, 2% SDS, 100 mm Tris-Cl, pH 6.8, at 65 °C for 30 min and loaded on a 15% acrylamide gel for separation by SDS-PAGE.
Protein Analyses
SDS-PAGE was performed using 20 μg of total protein for each lane and transferred in a semi-dry blotter to nitrocellulose membranes (Amersham Biosciences Protran 0.1 NC). The membrane was blocked 30 min with 3% dried milk in PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4) containing 0.1% (w/v) Tween 20 and incubated in primary antiserum; this solution was used as the diluent for both primary and secondary antibodies, for 1 h, respectively. PBS containing 0.1% (w/v) Tween 20 was used for washing membranes twice for 15 min each time. The secondary antibody, used at 1:10,000, was goat anti-rabbit conjugated to alkaline phosphatase. Antibodies directed against CF1 (1:15,000), Plastocyanin (1:5,000), OEE1 (1:5,000), PsaF (1:4000, J. D. Rochaix), D1 (1:1,000, Agrisera), Lhcb2 (1:5,000, Agrisera), PSAK (1:2,000, M. Hippler), CP29/Lchb4 (1:4000, F. A. Wollman), Lhca3 (1:8000, M. Hippler), LHCSR3 (1:1000, M. Hippler), CRD1 (1:1,000), and Cyt c6 (1:2,000) and HPPD (affinity purified against Arabidopsis HPPD, kindly provided by M. Matringe) (1:1,000) were used.
Quantitative Metal, Phosphorus and Sulfur Content Analysis
1 × 108 Chlamydomonas cells of a culture at a density of 3–5 × 106 cells/ml were collected by centrifugation at 2450 × g for 3 min in a 50-ml Falcon tube. The cells were washed three times in 1 mm Na2-EDTA, pH 8 (to remove cell surface-associated metals), and once in Milli-Q water. The cell pellet, after removing the water, was overlaid with 286 μl of 70% nitric acid and digested at room temperature for 24 h at 65 °C for about 2 h before being diluted to a final nitric acid concentration of 2% (v/v) with Milli-Q water. Aliquots of fresh or spent culture medium were treated with nitric acid to a final concentration of 2% (v/v). Metal, sulfur, and phosphorous contents were determined by inductively coupled plasma mass spectrometry on an Agilent 8800 Triple Quadrupole ICP-MS instrument, in comparison to an environmental calibration standard (Agilent 5183–4688), a sulfur (Inorganic Ventures CGS1), and phosphorus (Inorganic Ventures CGP1) standard, using 89Y (the 89 isotope of the chemical element Yttrium) as an internal standard (Inorganic Ventures MSY-100PPM). The levels of all analytes were determined in MS/MS mode, where 63Cu were measured directly using helium in the collision reaction cell, whereas 56Fe was directly determined using H2 as a cell gas. The average of 4 technical replicate measurements was used for each individual biological sample, the average variation between the technical replicate measurements was 1.1% for all analytes and never exceeded 5% for an individual sample. Triplicate samples (independent cultures) were used to determine the variation in between cultures, average, and standard deviation between these replicates are depicted in figures.
Fluorescence Measurements
Chlorophyll fluorescence emission spectra at 77 K were recorded with a TECANXP-HP8000 using intact cells diluted to a concentration of 5 μg of chlorophyll/ml in 30% glycerol, 5 mm Hepes, pH 7.5, and 10 mm EDTA and snap frozen in liquid nitrogen in a customized 96-well plate. The samples were excited at 435 nm (bandwidth 5 nm), and emission was monitored between 640 and 760 nm (bandwidth 5 nm). The gain was manually adjusted to 150, the Z-position was 22,000 μm. The signals obtained were processed with the Adjacent-Average function (OriginLab) and normalized to the value obtained at a wavelength of 685 nm.
Plastoquinone-9 and Tocochromanol Analyses
Chlamydomonas cells were grown to a density of 4–8 × 106 cells × ml−1 and 50–100 ml were collected by centrifugation (5 min at 2450 × g, 4 °C). For normalization, 1 ml of each culture was used for chlorophyll determination according to Ref. 58. For plastoquinone-9 and tocochromanol analyses, frozen cell pellets were resuspended in 500 μl of 95% (v/v) ethanol, spiked with 50 μl of 121 μm ubiquinone-10 (6.05 nmol) as an internal standard, and homogenized in a 5-ml Pyrex tissue grinder. The grinder was then rinsed with 500 μl of 95% (v/v) ethanol, and the wash was combined to the initial extract. Insoluble material was removed by centrifugation (5 min at 18,000 × g) and the supernatants were immediately analyzed by HPLC as previously described (60).
Author Contributions
D. S. and S. S. M. designed the study. D. S. designed and constructed vectors for conditional knock-down and performed hypoxia experiments. D. S. and C. A. L. performed copper deficiency and high light experiments. S. S. performed and analyzed ICP-MS measurements. A. F. and G. J. B. performed the tocopherol and plastoquinone experiments. D. S. and S. S. M. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Prof. Michel Matringe for the Arabidopsis thaliana HPPD immunopurifed antibody and Dr. Madeli Castruita for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM42143 (to S. M.) and National Science Foundation Grant MCB-1148968 (to G. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Cyt
- cytochrome
- CRR1
- copper response regulator 1
- HPPD
- 4-hydroxyphenylpyruvate dioxygenase
- Chl
- chlorophyll
- amiRNA
- artifical micro RNA
- qRT
- quantitative RT
- PC-8
- plastochromanol-8
- TAP
- Tris acetate/phosphate
- PQ-9
- plastoquinone-9
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- PSAK
- photosystem I subunit K.
References
- 1. Williams R. J. (2003) Metallo-enzyme catalysis. Chem. Commun. 2003, 1109–1113 [DOI] [PubMed] [Google Scholar]
- 2. Marschner H., and Marschner P. (2012) Marschner's Mineral Nutrition of Higher Plants, 3rd Ed., Elsevier/Academic Press, London [Google Scholar]
- 3. Merchant S. S., and Helmann J. D. (2012) Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60, 91–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Merchant S., Hill K., Quinn J., and Li H. H. (1995) Coordinate, copper-responsive expression of Cyt-c6, coproporphyrinogen oxidase and a copper uptake system. J. Cell. Biochem. 59, 241–241 [Google Scholar]
- 5. Quinn J. M., Barraco P., Eriksson M., and Merchant S. (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275, 6080–6089 [DOI] [PubMed] [Google Scholar]
- 6. Kropat J., Tottey S., Birkenbihl R. P., Depège N., Huijser P., and Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., and Shikanai T. (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernal M., Casero D., Singh V., Wilson G. T., Grande A., Yang H., Dodani S. C., Pellegrini M., Huijser P., Connolly E. L., Merchant S. S., and Krämer U. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24, 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Page M. D., Kropat J., Hamel P. P., and Merchant S. S. (2009) Two Chlamydomonas CTR copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21, 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peñarrubia L., Andrés-Colás N., Moreno J., and Puig S. (2010) Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J. Biol. Inorg. Chem. 15, 29–36 [DOI] [PubMed] [Google Scholar]
- 11. Castruita M., Casero D., Karpowicz S. J., Kropat J., Vieler A., Hsieh S. I., Yan W., Cokus S., Loo J. A., Benning C., Pellegrini M., and Merchant S. S. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moseley J., Quinn J., Eriksson M., and Merchant S. (2000) The CRD1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moseley J. L., Page M. D., Alder N. P., Eriksson M., Quinn J., Soto F., Theg S. M., Hippler M., and Merchant S. (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Albus C. A., Salinas A., Czarnecki O., Kahlau S., Rothbart M., Thiele W., Lein W., Bock R., Grimm B., and Schöttler M. A. (2012) LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 160, 1923–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hollingshead S., Kopecná J., Jackson P. J., Canniffe D. P., Davison P. A., Dickman M. J., Sobotka R., and Hunter C. N. (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287, 27823–27833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bollivar D., Braumann I., Berendt K., Gough S. P., and Hansson M. (2014) The Ycf54 protein is part of the membrane component of Mg-protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L.). FEBS J. 281, 2377–2386 [DOI] [PubMed] [Google Scholar]
- 17. Moran G. R. (2005) 4-Hydroxyphenylpyruvate dioxygenase. Arch. Biochem. Biophys. 433, 117–128 [DOI] [PubMed] [Google Scholar]
- 18. Maeda H., Sakuragi Y., Bryant D. A., and Dellapenna D. (2005) Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiol 138, 1422–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krieger-Liszkay A., and Trebst A. (2006) Tocopherol is the scavenger of singlet oxygen produced by the triplet states of chlorophyll in the PSII reaction centre. J. Exp. Bot. 57, 1677–1684 [DOI] [PubMed] [Google Scholar]
- 20. Gill S. S., and Tuteja N. (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930 [DOI] [PubMed] [Google Scholar]
- 21. Hussain N., Irshad F., Jabeen Z., Shamsi I. H., Li Z., and Jiang L. (2013) Biosynthesis, structural, and functional attributes of tocopherols in planta: past, present, and future perspectives. J. Agric. Food Chem. 61, 6137–6149 [DOI] [PubMed] [Google Scholar]
- 22. Trebst A., Depka B., and Holländer-Czytko H. (2002) A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 516, 156–160 [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi N., and DellaPenna D. (2008) Tocopherol metabolism, oxidation and recycling under high light stress in Arabidopsis. Plant J. 55, 607–618 [DOI] [PubMed] [Google Scholar]
- 24. Li Z., Keasling J. D., and Niyogi K. K. (2012) Overlapping photoprotective function of vitamin E and carotenoids in Chlamydomonas. Plant Physiol. 158, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zingg J. M. (2015) Vitamin E: a role in signal transduction. Annu. Rev. Nutr. 35, 135–173 [DOI] [PubMed] [Google Scholar]
- 26. Schmollinger S., Strenkert D., and Schroda M. (2010) An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr. Genet. 56, 383–389 [DOI] [PubMed] [Google Scholar]
- 27. Formighieri C., Ceol M., Bonente G., Rochaix J. D., and Bassi R. (2012) Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5, 1242–1262 [DOI] [PubMed] [Google Scholar]
- 28. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., and Loo J. A. (2013) The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hill K. L., Li H. H., Singer J., and Merchant S. (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6 analysis of the kinetics and metal specificity of its copper-Responsive expression. J. Biol. Chem. 266, 15060–15067 [PubMed] [Google Scholar]
- 30. Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., and Hippler M. (2002) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wollman F. A., and Delepelaire P. (1984) Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J. Cell Biol. 98, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wollman F. A., and Bennoun P. (1982) A new chlorophyll-protein complex related to photosystem-I in Chlamydomonas reinhardii. Biochim. Biophys. Acta 680, 352–360 [Google Scholar]
- 33. Urzica E. I., Casero D., Yamasaki H., Hsieh S. I., Adler L. N., Karpowicz S. J., Blaby-Haas C. E., Clarke S. G., Loo J. A., Pellegrini M., and Merchant S. S. (2012) Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Allorent G., Tokutsu R., Roach T., Peers G., Cardol P., Girard-Bascou J., Seigneurin-Berny D., Petroutsos D., Kuntz M., Breyton C., Franck F., Wollman F. A., Niyogi K. K., Krieger-Liszkay A., Minagawa J., and Finazzi G. (2013) A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonente G., Ballottari M., Truong T. B., Morosinotto T., Ahn T. K., Fleming G. R., Niyogi K. K., and Bassi R. (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLos Biol. 9, e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergner S. V., Scholz M., Trompelt K., Barth J., Gäbelein P., Steinbeck J., Xue H., Clowez S., Fucile G., Goldschmidt-Clermont M., Fufezan C., and Hippler M. (2015) STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol. 168, 615–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ballottari M., Truong T. B., De Re E., Erickson E., Stella G. R., Fleming G. R., Bassi R., and Niyogi K. K. (2016) Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 291, 7334–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Correa-Galvis V., Redekop P., Guan K., Griess A., Truong T. B., Wakao S., Niyogi K. K., and Jahns P. (2016) Photosystem II subunit PsbS is involved in the induction of LHCSR-dependent energy dissipation in Chlamydomonas reinhardtii. J. Biol. Chem. 10.1074/jbc.116.737312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petroutsos D., Busch A., Janssen I., Trompelt K., Bergner S. V., Weinl S., Holtkamp M., Karst U., Kudla J., and Hippler M. (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23, 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hemschemeier A., Casero D., Liu B., Benning C., Pellegrini M., Happe T., and Merchant S. S. (2013) Copper response regulator 1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell 25, 3186–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kropat J., Gallaher S. D., Urzica E. I., Nakamoto S. S., Strenkert D., Tottey S., Mason A. Z., and Merchant S. S. (2015) Copper economy in Chlamydomonas: prioritized allocation and reallocation of copper to respiration vs. photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 2644–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huertas M. J., López-Maury L., Giner-Lamia J., Sánchez-Riego A. M., and Florencio F. J. (2014) Metals in cyanobacteria: analysis of the copper, nickel, cobalt and arsenic homeostasis mechanisms. Life 4, 865–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wood P. M. (1978) Interchangeable copper and iron proteins in algal photosynthesis: studies on plastocyanin and cytochrome c552 in Chlamydomonas. Eur. J. Biochem. 87, 9–19 [DOI] [PubMed] [Google Scholar]
- 44. Sandmann G., Reck H., Kessler E., and Boger P. (1983) Distribution of plastocyanin and soluble plastidic cytochrome-c in various classes of algae. Arch. Microbiol. 134, 23–27 [Google Scholar]
- 45. Merchant S., and Bogorad L. (1986) Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell. Biol. 6, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kruk J., Szymańska R., Cela J., and Munne-Bosch S. (2014) Plastochromanol-8: fifty years of research. Phytochemistry 108, 9–16 [DOI] [PubMed] [Google Scholar]
- 47. Eugeni Piller L., Abraham M., Dörmann P., Kessler F., and Besagni C. (2012) Plastid lipid droplets at the crossroads of prenylquinone metabolism. J. Exp. Bot. 63, 1609–1618 [DOI] [PubMed] [Google Scholar]
- 48. Fatihi A., Latimer S., Schmollinger S., Block A., Dussault P. H., Vermaas W. F., Merchant S. S., and Basset G. J. (2015) A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of vitamin K1 in Synechocystis and Arabidopsis. Plant Cell 27, 1730–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinis J., Glauser G., Valimareanu S., and Kessler F. (2013) A chloroplast ABC1-like kinase regulates vitamin E metabolism in Arabidopsis. Plant Physiol 162, 652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., and Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 51. Brzezowski P., Richter A. S., and Grimm B. (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochem. Biophys. Acta 1847, 968–985 [DOI] [PubMed] [Google Scholar]
- 52. Pinnola A., Cazzaniga S., Alboresi A., Nevo R., Levin-Zaidman S., Reich Z., and Bassi R. (2015) Light-harvesting complex stress-related proteins catalyze excess energy dissipation in both photosystems of Physcomitrella patens. Plant Cell 27, 3213–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amunts A., Toporik H., Borovikova A., and Nelson N. (2010) Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., and Baulcombe D. (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58, 165–174 [DOI] [PubMed] [Google Scholar]
- 55. Kindle K. L. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sommer F., Kropat J., Malasarn D., Grossoehme N. E., Chen X., Giedroc D. P., and Merchant S. S. (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22, 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., and Malasarn D. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Porra R. J., Thompson W. A., and Kriedemann P. E. (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-B extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 59. Strenkert D., Schmollinger S., Sommer F., Schulz-Raffelt M., and Schroda M. (2011) Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23, 2285–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Block A., Fristedt R., Rogers S., Kumar J., Barnes B., Barnes J., Elowsky C. G., Wamboldt Y., Mackenzie S. A., Redding K., Merchant S. S., and Basset G. J. (2013) Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J. Biol. Chem. 288, 27594–27606 [DOI] [PMC free article] [PubMed] [Google Scholar]