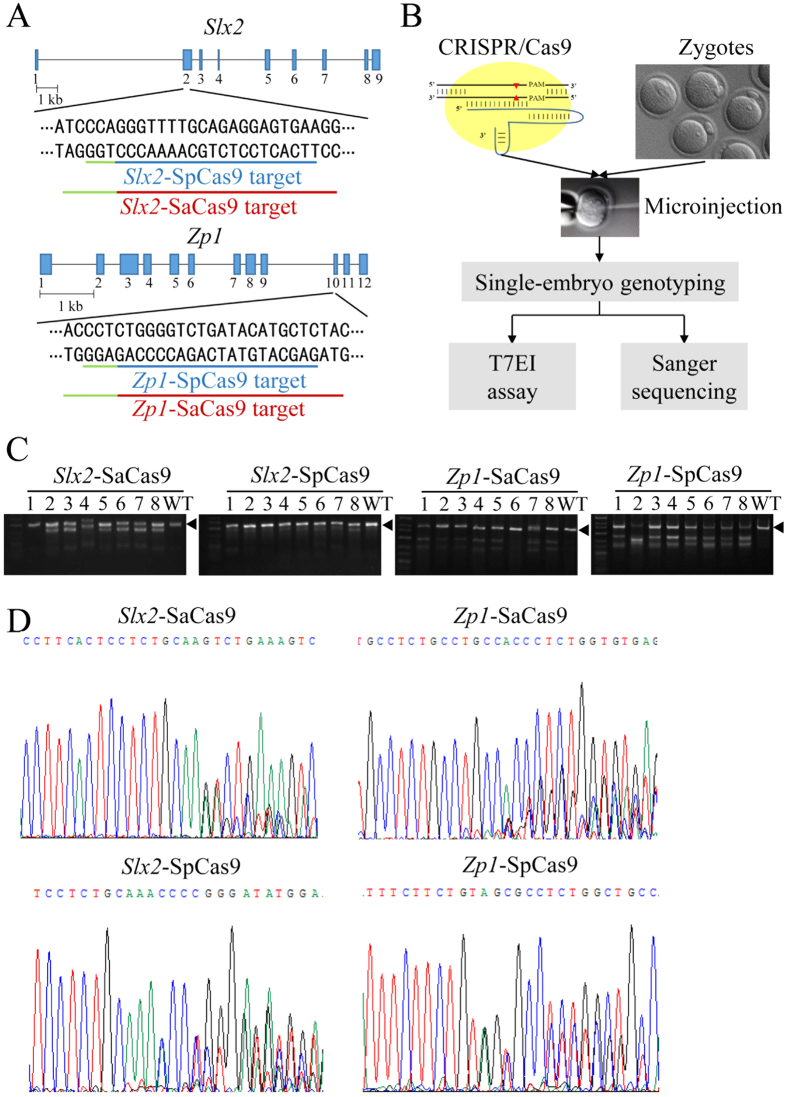
Figure 1. SaCas9 can efficiently cleave target genes in mouse embryos.
(A) Schematic representations of the Slx2 and Zp1 target sites. Here, SaCas9 and SpCas9 target the same regions of Slx2 and Zp1, their target sequences are underlined respectively in red and blue. PAM sequences are underlined in green. (B) A flow chart indicating the strategy to determine the cleavage efficiencies of SaCas9. SpCas9 was used for comparsion. Cas9 mRNAs and their respective gRNAs were injected into 0.5-day old zygotes. After 48 hours, a portion of the injected embryos were in vitro cultured for the T7 endonuclease I (T7EI) assay and Sanger sequencing. (C) The regions spanning the target sites of the Slx2 and Zp1 loci were PCR amplified. The PCR products were then used for the T7EI assay. Black arrowheads indicate PCR fragments that contain no mismatch. WT, wild-type. (D) The PCR products from (C) were also examined by Sanger sequencing. Representative sequencing results of SaCas9 or SpCas9-cleaved embryos are presented here. The presences of multiple peaks near the PAM sequences indicate sites of cleavage and NHEJ-mediated repair.