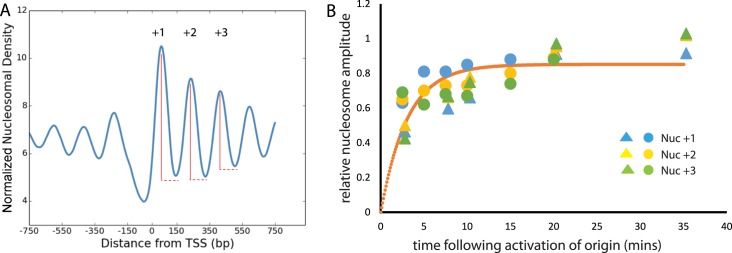
Figure 3.
Kinetics of nucleosome organization. The depth of the oscillation in nucleosomal read depth was determined for the +1, +2 and +3 nucleosomes as indicated in (A). The oscillation depth in nascent chromatin at nuc +1 (blue), nuc +2 (yellow), nuc +3 (green) was then expressed as a fraction of that observed in the input chromatin for two repeats of an EdU time course at the time points indicated (B). The time values were calculated based on the distribution of replicated fragments observed at origins multiplied by the rate of elongation for DNA polymerase, 1.6 kb/min (17). Time points for two biological repeats are shown as circles and triangles. A fit to the first order rate equation, y = Ae−(kt) is shown (orange) which allows estimation of the half time for nucleosome positioning as 2.1 min. The residual, R2, for this fit is 0.48.