Figure 4.
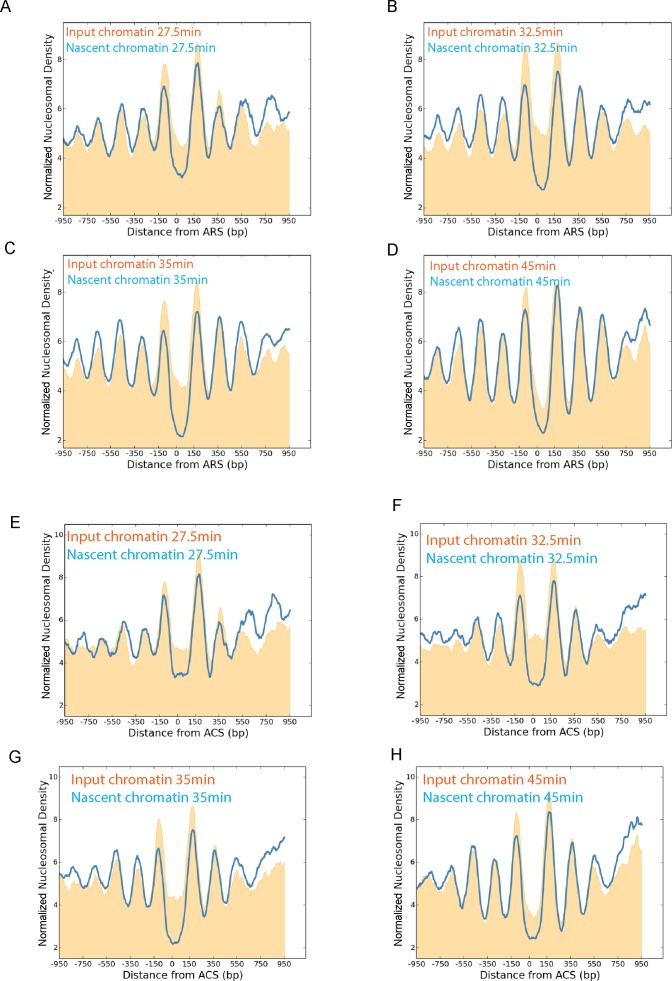
Chromatin maturation at origins of DNA replication. Chromatin from the EdU enrichment time course was plotted with respect to 205 replication origins (46). Nascent (blue) and input chromatin (orange) are plotted 27.5 (A), 32.5 (B), 35 (C) and 45 min (D) following release from G1 arrest. The +2 and +3 Nucleosomes are significantly ordered at the first time point and this improves over the following minutes. As many replication origins are located adjacent to transcribed genes, the same analysis was performed with 127 replication origins for which no TSS was present within 500 bp of the origin (E–H). Nucleosomes are not as precisely aligned to TSS-free origins in comparison to all origins (Compare input chromatin A–D to that of E–H). In particular the nucleosome depleted region at origins is poorly defined in early S-phase. Organization of the +2 and +3 nucleosomes at replication origins with no adjacent TSS mature at a similar rate to that observed at all origins.