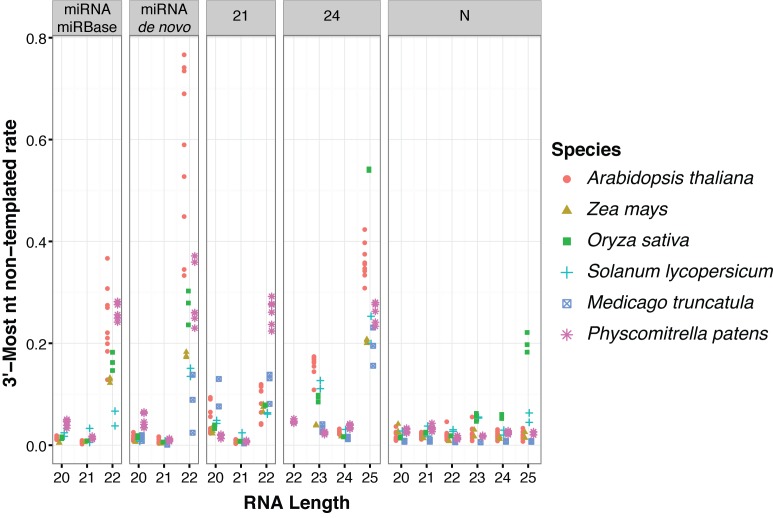
Figure 3.
‘Off-sized’ miRNAs and siRNAs have high rates of 3′ end non-templated nucleotides in several plant species. Mismatch rates for reference-aligned sRNA-seq reads of the indicated species, clusters and sizes. 21: siRNA loci dominated by 21 nt RNAs. 24: siRNA loci dominated by 24 nt siRNAs (presumed het-siRNA loci). N: loci where less than 80% of reads were between 20–24 nts, and are thus presumed to not be miRNA or siRNA loci. miRNA-miRBase: High-confidence MIRNA loci from miRBase version 21. miRNA-de novo: MIRNA loci annotated by automated software. Note that most de novo annotated MIRNA loci overlapped prior miRBase annotations. Libraries that were analyzed are listed in Supplementary Table S1.