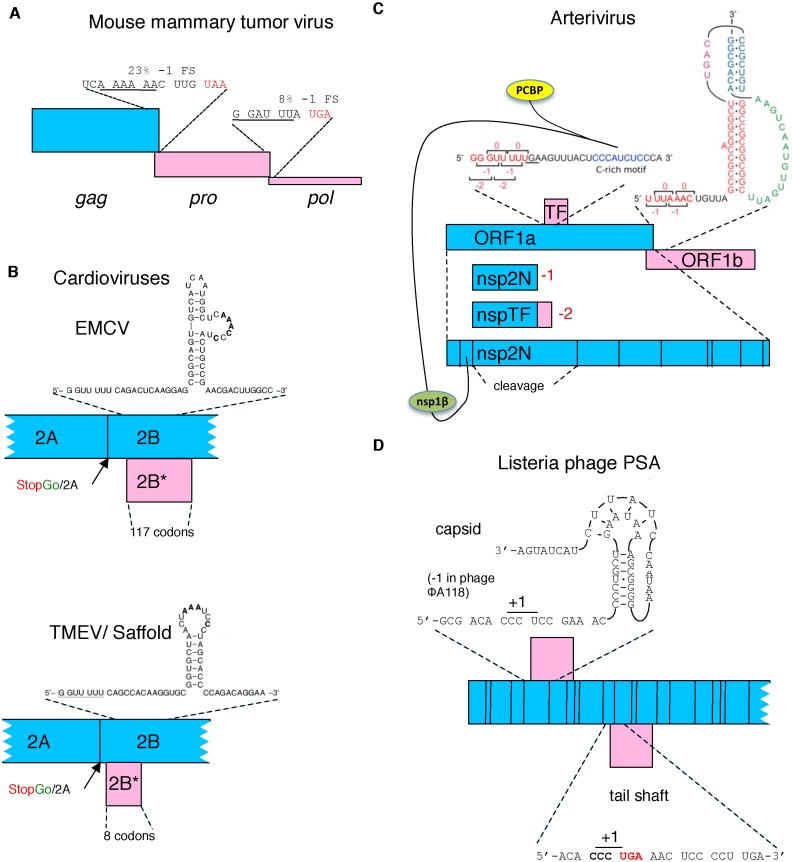
Figure 3.
Schematic representation of several viral frameshifting cassettes. (A) Retroviral frameshifting with the thickness of the pro and pol ORFs (pink) reflecting frameshift efficiency and the proportion of ribosomes that decode them with respect to zero frame gag (blue). (B) The −1 ribosomal frameshift site in two cardioviruses that yields a transframe encoded protein (pink). The proximity of the Theiler's murine encephalomyelitis virus (TMEV) shift site to the StopGo is evident and the relevant amino acid sequence is shown in Table 2. (C) In arteriviruses, −1 ribosomal frameshifting near the end of a long 5′ gene, ORF1a, leads a proportion of ribosomes to continue synthesis by decoding a second and also long gene (ORF1b), with the products of both ORFs specifying non-structural polyproteins. At least eight shorter 3′ ORFs encode structural proteins. −2 ribosomal frameshifting in a central region of ORF1a causes some ribosomes to access the wholly overlapping +1 frame TF ORF to yield a C-terminal extension to nsp2 (the product liberated by proteolytic cleavage from that region of the polyprotein encoded by ORF1a). This frameshifting is stimulated in trans by virus-encoded nsp1β in complex with poly(C) binding protein (PCBP). (D) Listeria phage PSA. Capsid and tail shaft encoding genes utilize +1 ribosomal frameshifting on a proline codon just 5′ of either a stimulatory pseudoknot (capsid) or stop codon (tail shaft).