Figure 5.
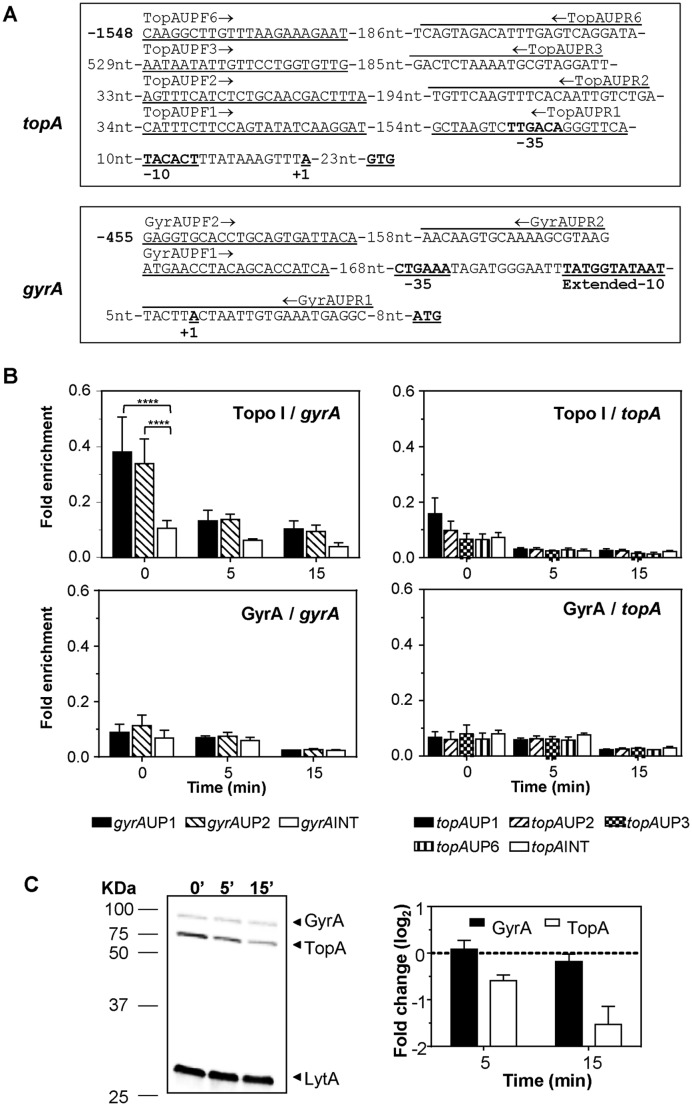
Recruitment of Topo I and GyrA to topA and gyrA upstream sequences. (A) Sequence of the topA and gyrA upstream regions and localization of the oligonucleotides used in the qRT-PCR experiments. The −35 and −10 boxes, and the nucleotide in which transcription is initiated (+1) are indicated. (B) Determination of Topo I or gyrase binding on upstream topA or gyrA regions by ChIP-qRT-PCR. Exponentially growing cells were subjected to chromatin immunoprecipitation (ChIP) using anti-Topo I or anti-GyrA antibodies, and the pulled-down DNA was subsequently analyzed by qPCR. The graphs show pulldown efficiency (ChIP-DNA/input DNA) for each primer pair. Values are the average ±SD of three independent replicates. ****P < 0.0001. (C) Western blot analysis of GyrA and Topo I performed as described in the legend of Figure 4.