Fig. 2.
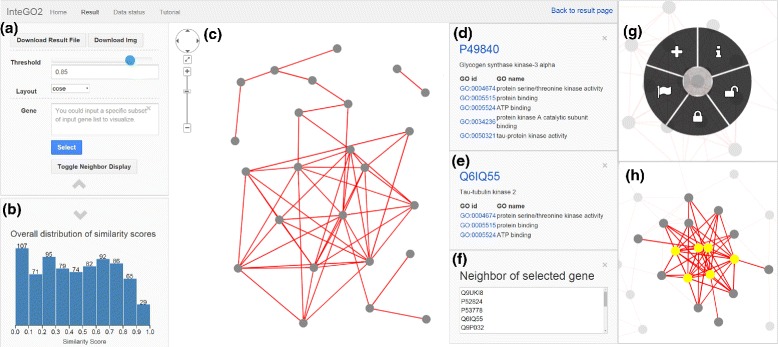
The visualization interface of I n t e G O2 to explore gene functional similarities based on GO. The network is shown in panel (c), in which a node represents a gene, and an edge indicates that the similarity score between the two corresponding genes is larger than an edge similarity threshold, which can be changed in panel (a). Edge similarity scores distribution shown in panel (b) helps users to choose an appropriate threshold. The gene information panel (d) and (e) show the recently chosen genes and current gene respectively. Panel f shows the neighbors of the recently chosen genes. The node operation panel (g) allows users to flag, lock or unlock a gene. The selected subnetwork is shown in (h)
