Abstract
Background
Recently, structured sparse canonical correlation analysis (SCCA) has received increased attention in brain imaging genetics studies. It can identify bi-multivariate imaging genetic associations as well as select relevant features with desired structure information. These SCCA methods either use the fused lasso regularizer to induce the smoothness between ordered features, or use the signed pairwise difference which is dependent on the estimated sign of sample correlation. Besides, several other structured SCCA models use the group lasso or graph fused lasso to encourage group structure, but they require the structure/group information provided in advance which sometimes is not available.
Results
We propose a new structured SCCA model, which employs the graph OSCAR (GOSCAR) regularizer to encourage those highly correlated features to have similar or equal canonical weights. Our GOSCAR based SCCA has two advantages: 1) It does not require to pre-define the sign of the sample correlation, and thus could reduce the estimation bias. 2) It could pull those highly correlated features together no matter whether they are positively or negatively correlated. We evaluate our method using both synthetic data and real data. Using the 191 ROI measurements of amyloid imaging data, and 58 genetic markers within the APOE gene, our method identifies a strong association between APOE SNP rs429358 and the amyloid burden measure in the frontal region. In addition, the estimated canonical weights present a clear pattern which is preferable for further investigation.
Conclusions
Our proposed method shows better or comparable performance on the synthetic data in terms of the estimated correlations and canonical loadings. It has successfully identified an important association between an Alzheimer’s disease risk SNP rs429358 and the amyloid burden measure in the frontal region.
Keywords: Brain imaging genetics, Canonical correlation analysis, Structured sparse model, Machine learning
Background
In recent years, the bi-multivariate analyses techniques [1], especially the sparse canonical correlation analysis (SCCA) [2–8], have been widely used in brain imaging genetics studies. These methods are powerful in identifying bi-multivariate associations between genetic biomarkers, e.g., single nucleotide polymorphisms (SNPs), and the imaging factors such as the quantitative traits (QTs).
Witten et al. [3, 9] first employed the penalized matrix decomposition (PMD) technique to handle the SCCA problem which had a closed form solution. This SCCA imposed the ℓ1-norm into the traditional CCA model to induce sparsity. Since the ℓ1-norm only randomly chose one of those correlated features, it performed poorly in finding structure information which usually existed in biology data. Witten et al. [3, 9] also implemented the fused lasso based SCCA which penalized two adjacent features orderly. This SCCA could capture some structure information but it demanded the features be ordered. As a result, a lot of structured SCCA approaches arose. Lin et al. [7] imposed the group lasso regularizer to the SCCA model which could make use of the non-overlapping group information. Chen et al. [10] proposed a structure-constrained SCCA (ssCCA) which used a graph-guided fused ℓ2-norm penalty for one canonical loading according to features’ biology relationships. Du et al. [8] proposed a structure-aware SCCA (S2CCA) to identify group-level bi-multivariate associations, which combined both the covariance matrix information and the prior group information by the group lasso regularizer. These structured SCCA methods, on one hand, can generate a good result when the prior knowledge is well fitted to the hidden structure within the data. On the other hand, they become unapproachable when the prior knowledge is incomplete or not available. Moreover, it is hard to precisely capture the prior knowledge in real world biomedical studies.
To facilitate structural learning via grouping the weights of highly correlated features, the graph theory were widely utilized in sparse regression analysis [11–13]. Recently, we notice that the graph theory has also been employed to address the grouping issue in SCCA. Let each graph vertex and each feature has a one-to-one correspondence relationship, and ρij be the sample correlation between features i and j. Chen et al. [4, 5] proposed a network-structured SCCA (NS-SCCA) which used the ℓ1-norm of |ρij|(ui−sign(ρij)uj) to pull those positively correlated features together, and fused those negatively correlated features to the opposite direction. The knowledge-guided SCCA (KG-SCCA) [14] was an extension of both NS-SCCA [4, 5] and S2CCA [8]. It used ℓ2-norm of for one canonical loading, similar to what Chen proposed, and employed the ℓ2,1-norm penalty for another canonical loading. Both NS-SCCA and KG-SCCA could be used as a group-pursuit method if the prior knowledge was not available. However, one limitation of both models is that they depend on the sign of pairwise sample correlation to recover the structure pattern. This probably incur undesirable bias since the sign of the correlations could be wrongly estimated due to possible graph misspecification caused by noise [13].
To address the issues above, we propose a novel structured SCCA which neither requires to specify prior knowledge, nor to specify the sign of sample correlations. It will also work well if the prior knowledge is provided. The GOSC-SCCA, named from Graph Octagonal Selection and Clustering algorithm for Sparse Canonical Correlation Analysis, is inspired by the outstanding feature grouping ability of octagonal selection and clustering algorithm for regression (OSCAR) [11] regularizer and graph OSCAR (GOSCAR) [13] regularizer in regression task. Our contributions can be summarized as follows 1) GOSC-SCCA could pull those highly correlated features together when no prior knowledge is provided. While those positively correlated features will be encouraged to have similar weights, those negatively correlated ones will also be encouraged to have similar weights but with different signs. 2) Our GOSC-SCCA could reduce the estimation bias given no requirement for specifying the sign of sample correlation. 3) We provide a theoretical quantitative description for the grouping effect of GOSC-SCCA. We use both synthetic data and real imaging genetic data to evaluate GOSC-SCCA. The experimental results show that our method is better than or comparable to those state-of-the-art methods, i.e., L1-SCCA, FL-SCCA [3] and KG-SCCA [14], in identifying stronger imaging genetic correlations and more accurate and cleaner canonical loadings pattern. Note that the PMA software package were used to implement the L1-SCCA (SCCA with lasso penalty) and FL-SCCA (SCCA with fused lasso penalty) methods. Please refer to http://cran.r-project.org/web/packages/PMA/ for more details.
Methods
We denote a vector as a boldface lowercase letter, and denote a matrix as a boldface uppercase letter. mi indicates the i-th row of matrix M=(mij). Matrices and denote two separate datasets collected from the same population. Imposing lasso into a traditional CCA model [15], the L1-SCCA model is formulated as follows [3, 9]:
| 1 |
where ||u||1≤c1 and ||v||1≤c2 are sparsity penalties controlling the complexity of the SCCA model. The fused lasso [2–4, 9] can also be used instead of lasso. In order to make the problem be convex, the equal sign is usually replaced by less-than-equal sign, i.e. [3].
The graph OSCAR regularization
The OSCAR regularizer is firstly introduced by Bondell et al. [11], which has been proved to have the ability of grouping features automatically by encouraging those highly correlated features to have similar weights. Formally, the OSCAR penalty is defined as follows,
| 2 |
Note that this penalty is applied to each feature pair.
To make OSCAR be more flexible, Yang et al. [13] introduce the GOSCAR,
| 3 |
where Eu and Ev are the edge sets of the u-related and v-related graphs, respectively. Obviously, the GOSCAR will reduce to OSCAR when both graphs are complete [13].
Applying , the GOSCAR regularizer takes the following form,
| 4 |
The GOSC-SCCA model
Since the grouping effect is also an important consideration in SCCA learning, we propose to expand L1-SCCA to GOSC-SCCA by imposing GOSCAR instead of L1 only as follows.
| 5 |
where (c1,c2,c3,c4) are parameters and they could control the solution path of the canonical loadings. Since the S2CCA [8] has proved that the covariance matrix information could help improve the prediction ability, we also use and other than .
As a structured sparse model, GOSC-SCCA will encourage if the i-th feature and the j-th feature are highly correlated. We will give a quantitative description for this later.
The proposed algorithm
We can write the objective function into unconstrained formulation via the Lagrange multiplier method, i.e.
| 6 |
where (λ1,λ2,β1,β2) are tuning parameters, and they have a one-to-one correspondence to parameters (c1,c2,c3,c4) in GOSC-SCCA model [4].
Taking the derivative regarding u and v respectively, and letting them be zero, we obtain,
| 7 |
| 8 |
where Λ1 is a diagonal matrix with the k1-th element as , and Λ2 with the k2-th element as ; L1 is the Laplacian matrix which can be obtained from L1=D1−W1; is a matrix which is from . L2 and have the same entries as L1 and separately based on v.
In the initialization, both W1 and have the same entry with each element as except the diagonal elements. But W1 and become different after each iteration, i.e.,
| 9 |
If ||ui−uj||1=0, the corresponding element in matrix W1 will not exist. So we regularize it as (ζ is a very small positive number) when ||ui−uj||1=0. We also approximate ||ui||1=0 with for Λ1. Then the objective function regarding u is . It is easy to prove that will reduce to problem (6) regarding u when ζ→0. The cases of ||vi||1=0 and ||vi−vj||1=0 can be addressed using a similar regularization method.
D1 is a diagonal matrix and its i-th diagonal element is obtained by summing the i-th row of W1, i.e. . The diagonal matrix is also obtained from . Likewise, we can calculate W2, , D2 and by the same method in terms of v.
Then according to Eqs. (7-8), we can obtain the solution to our problem with respect to u and v separately.
| 10 |
| 11 |

We observe that L1, and Λ1 depend on u which is an unknown variable, and v is also unknown which is used to calculate L2, and Λ2. Thus we propose an effective iterative algorithm to solve this problem. We first fix v to solve u; and then fix u to solve v.
Algorithm 1 exhibits the pseudo code of the proposed GOSC-SCCA algorithm. For the key calculation steps, i.e., Step 5 and Step 10, we solve a system of linear equations with quadratic complexity other than computing the matrix inverse with cubic complexity. Thus the whole algorithm can work with desired efficiency. In addition, the algorithm is guaranteed to converge and we will prove this in the next subsection.
Convergence analysis
We first introduce the following lemma.
Lemma 1
For any two nonzero real numbers and u, we have
| 12 |
Proof
Given the lemma in [16], we have for any two nonzero vectors. We also have and ||u||1=||u||2 for any two nonzero real numbers, which completes the proof. □
Based on Lemma 1, we have
| 13 |
| 14 |
when , , and are nonzero.
We now have the following theorem regarding GOSC-SCCA algorithm.
Theorem 1
The objective function value of GOSC-SCCA will monotonically decrease in each iteration till the algorithm converges.
Proof
The proof consists of two parts.
(1) Part 1: From Step 3 to Step 7 in Algorithm 1, u is the only unknown variable to be solved. The objective function (6) can be equivalently transferred to
According to Step 5 we have
where is the updated u.
It is known that if L is the laplacian matrix [17]. Similarly, . Then according to Eq. (9), we obtain
| 15 |
We first multiply 2λ1 on both sides of Eq. (13) for each feature pair separately, and do the same to both sides of Eq. (14). After that, we multiply β1 on both sides of Eq. (12). Finally, by summing all these inequations together to both sides of Eq. (15) accordingly, we arrive at
Let , , we have
| 16 |
Therefore, GOSC-SCCA will decrease the objective function in each iteration, i.e., .
(2) Part 2: From Step 8 to Step 12, the only unknown variable is v. Similarly, we can arrive at
| 17 |
Thus GOSC-SCCA also decreases the objective function in each iteration during the second phase, i.e., .
Based on the analysis above, we easily have according to the transitive property of inequalities. Therefore, the objective value monotonically decreases in each iteration. Note that the CCA objective ranges from [-1,1], and both uTXTXu and vTYTYv are constrained to be 1. Thus the −uTXTYv is lower bounded by -1, and so Eq. (6) is lower bounded by –1. In addition, Eqs. (16–17) imply that the KKT condition is satisfied. Therefore, the GOSC-SCCA algorithm will converge to a local optimum. □
Based on the convergence analysis, to facilitate the GOSC-SCCA algorithm, we set the stopping criterion of Algorithm 1 as max{|δ|∣δ∈(ut+1−ut)}≤τ and max{|δ|∣δ∈(vt+1−vt)}≤τ, where τ is a predefined estimation error. Here we set τ=10−5 empirically from the experiments.
The grouping effect of GOSC-SCCA
For the structured sparse learning in high-dimensional situation, the automatic feature grouping property is of great importance [18]. In regression analysis, Zou and Hastie [18] have suggested that a regressor behaviors grouping effect when it can set those regression coefficients of the same group to similar weights. This is also the case for structured SCCA methods. So, it is important and meaningful to investigate the theoretical boundary of the grouping effect.
We have the following theorem in terms of GOSC-SCCA.
Theorem 2
Let X and Y be two data sets, and (λ,β,γ)be the pre-tuned parameters. Let be the solution to our SCCA problem of Eqs. (10–11). Suppose the i-th feature and j-th feature only link to each other on the graph, and are their optimal solutions, thus holds. The solutions to and satisfy
| 18 |
where ρij is the sample correlation between features i and j, and wi,j is the corresponding element in u-related matrix W1.
Proof
Let be the solution to our problem Eq. (6), we have the following equations after taking the partial derivative with respect to and , respectively.
We know that features i and uj are only linked to each other, thus Dii=Djj=Aij=wij for those intermediate matrices. Besides, we also know that , , and . Then according to the definition of L1, and Λ1, we can arrive at
| 19 |
Subtracting these two equations, we obtain
| 20 |
Then we take ℓ2-norm on both sides of Eq. (20), apply the triangle inequality, and use the equality ,
| 21 |
We have known that our problem implies , thus we arrive at
| 22 |
□
Now the upper bound for the canonical loadings v can also be obtained, i.e.
| 23 |
where ρij′ is the sample correlation between the i-th and j-th feature in v, and is the corresponding element in v-related matrix W2.
Theorem 2 provides a theoretical upper bound for the difference between the estimated coefficients of the i-th feature and j-th feature. It seems that this is not a tight enough bound. However our bound is slack since it does not bound much more the pairwise difference of features i and j if ρij≪1. This is desirable for two irrelevant features [19]. Suppose two features with very small correlation, i.e. ρij≪0, their coefficients do not need to be the same or similar. So we do not care about their coefficients’ pairwise difference, and will not set their pairwise difference a tight bound. This quantitative description for the grouping effect makes the GOSCAR penalty an ideal choice for structured SCCA.
Results
We compare GOSC-SCCA with several state-of-the-art SCCA and structured SCCA methods, including L1-SCCA [3], FL-SCCA [3], KG-SCCA [14]. We do not compare GOSC-SCCA with S2CCA [8], ssCCA [7] and CCA-SG (CCA Sparse Group) [10] since they require prior knowledge available in advance. We do not choose NS-SCCA [5] as benchmark either, due to the following two reasons. (1) NS-SCCA generates many intermediate variables during its iterative procedure. As the authors stated, NS-SCCA’s per-iteration complexity is linear in (p+|E|), and thus the complexity becomes O(p2) when it is in the group pursuit mode. (2) Its penalty term is similar to that of KG-SCCA which has been selected for comparison.
There are six parameters to be decided before using the GOSC-SCCA, thus it will take too much time by blindly tuning. We tune the parameters following two principles. On one hand, Chen and Liu [5] found out that the result is not very sensitive to γ1 and γ2. So we choose them from a small scope [0.1, 1, 10]. On the other hand, if the parameters are too small, the SCCA will reduce to CCA due to the subtle influence of the penalties. And, too large parameters will over-penalize the results. Therefore, we tune the rest of the parameters within the range of {10−3,10−2,10−1,100,101,102,103}. In this study, we conduct all the experiments using the nested 5-fold cross-validation strategy, and the parameters are only tuned from the training set. In order to save time, we only tune these parameters on the first run of the cross-validation. That is, the parameters are tuned when the first four folds are used as the training set. Then we directly use the tuned parameters for all the remaining experiments. All these methods use the same partition for cross-validation in the experiment.
Evaluation on synthetic data
We generate four synthetic datasets to investigate the performance of GOSC-SCCA and those benchmarks. Following [4, 5], these datasets are generated by four steps: 1) We predefine the structures and use them to create u and v respectively. 2) We create a latent vector z from N(0,In×n). 3) We create X with each where and Y with each where . 4) For the first group of nonzero features in u, we change half of their signs, and also change the signs of the corresponding data. Since the synthetic datasets are order-independent, this setup is equivalent to randomly change a portion of features’ signs in u. Now that we change the sign of both coefficients and the data simultaneously, we still have X′u′=Xu where X′ and u′ indicate the data and coefficients after the sign swap. We do the same on the Y side to make our simulation more challenging [13]. In addition, we set all four datasets with n=80, p=100 and q=120. They also have different correlation coefficients and different group structures. Therefore, the simulation is designed to cover a set of diverse cases for a fair comparison.
The estimated correlation coefficients of each method on four datasets are contained in Table 1. The best values and those are not significantly worsen than the best values are shown in bold. On the training results, we observe that GOSC-SCCA either estimates the largest correlation coefficients (Dataset 1 and Dataset 4), or is not significantly worse than the best method (Dataset 2 and Dataset 3). GOSC-SCCA also has the best average correlation coefficients. On the testing results, GOSC-SCCA also outperforms those benchmarks in terms of the average correlation coefficients, though KG-SCCA does not perform significantly worse than our method. For the overall average obtained across four datasets, GOSC-SCCA obtains the better correlation coefficients than the competing methods on both training set and testing set.
Table 1.
5-fold cross-validation results on synthetic data
| Training results | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methods | Dataset 1 | MEAN | Dataset 2 | MEAN | Dataset 3 | MEAN | Dataset 4 | MEAN | AVG. | ||||||||||||||||
| L1-SCCA | 0.52 | 0.56 | 0.52 | 0.53 | 0.51 | 0.53 | 0.25 | 0.29 | 0.16 | 0.20 | 0.23 | 0.23 | 0.56 | 0.24 | 0.57 | 0.53 | 0.52 | 0.48 | 0.46 | 0.50 | 0.53 | 0.48 | 0.35 | 0.46 | 0.43 |
| FL-SCCA | 0.52 | 0.60 | 0.52 | 0.53 | 0.50 | 0.53 | NaN | NaN | 0.17 | NaN | 0.23 | 0.08 | 0.63 | 0.43 | 0.56 | 0.55 | 0.55 | 0.54 | 0.51 | 0.56 | NaN | 0.53 | 0.40 | 0.40 | 0.39 |
| KG-SCCA | 0.52 | 0.55 | 0.52 | 0.53 | 0.53 | 0.53 | 0.25 | 0.29 | 0.15 | 0.20 | 0.22 | 0.22 | 0.56 | 0.24 | 0.43 | 0.52 | 0.52 | 0.45 | 0.51 | 0.56 | 0.48 | 0.52 | 0.40 | 0.49 | 0.42 |
| GOSC-SCCA | 0.57 | 0.62 | 0.57 | 0.59 | 0.63 | 0.60 | 0.26 | 0.30 | 0.15 | 0.21 | 0.17 | 0.22 | 0.64 | 0.31 | 0.42 | 0.61 | 0.59 | 0.51 | 0.51 | 0.56 | 0.55 | 0.54 | 0.41 | 0.52 | 0.46 |
| Testing results | |||||||||||||||||||||||||
| L1-SCCA | 0.57 | 0.43 | 0.58 | 0.49 | 0.59 | 0.53 | 0.00 | 0.21 | 0.32 | 0.17 | 0.08 | 0.16 | 0.36 | 0.20 | 0.37 | 0.49 | 0.46 | 0.38 | 0.45 | 0.29 | 0.20 | 0.40 | 0.67 | 0.40 | 0.37 |
| FL-SCCA | 0.56 | 0.38 | 0.57 | 0.49 | 0.59 | 0.52 | NaN | NaN | 0.48 | NaN | 0.08 | 0.11 | 0.30 | 0.80 | 0.36 | 0.51 | 0.41 | 0.47 | 0.55 | 0.30 | NaN | 0.46 | 0.72 | 0.40 | 0.38 |
| KG-SCCA | 0.56 | 0.43 | 0.57 | 0.49 | 0.58 | 0.53 | 0.00 | 0.21 | 0.31 | 0.18 | 0.07 | 0.15 | 0.37 | 0.20 | 0.45 | 0.50 | 0.45 | 0.39 | 0.52 | 0.29 | 0.34 | 0.46 | 0.71 | 0.46 | 0.38 |
| GOSC-SCCA | 0.73 | 0.39 | 0.68 | 0.56 | 0.45 | 0.56 | 0.02 | 0.09 | 0.57 | 0.20 | 0.38 | 0.25 | 0.23 | 0.18 | 0.43 | 0.44 | 0.43 | 0.34 | 0.53 | 0.31 | 0.31 | 0.36 | 0.72 | 0.45 | 0.40 |
The estimated correlation coefficients and their MEAN are shown. ’NaN’ means a method fails to estimate a pair of canonical loadings. ’0.00’ means a very small correlation coefficients. ’AVG.’ denotes the MEAN across all four datasets. The best values and those that are NOT significantly worse than the best ones (t-test with p-value smaller than 0.05) are shown in bold
Figure 1 shows the estimated canonical loadings of all four SCCA methods in a typical run. As we can see, L1-SCCA cannot accurately recover the true signals. For those coefficients with sign swapped, it fails to recognize them. The FL-SCCA slightly improves L1-SCCA’s performance but cannot identify those coefficients with sign changed either. Our GOSC-SCCA successfully groups those nonzero features together, and accurately recognizes the coefficients whose signs are changed. No matter what structures are within the dataset, GOSC-SCCA is able to estimate true signals which are very close to the ground truth. Although KG-SCCA also recognizes the coefficients with sign swapped, it is unable to recover every group of nonzero coefficients. For example, KG-SCCA misses two groups of nonzero features in terms of v for the second dataset. The results on synthetic datasets reveal that GOSC-SCCA can not only estimate stronger correlation coefficients than the competing methods, but also identifies more accurate and cleaner canonical loadings.
Fig. 1.
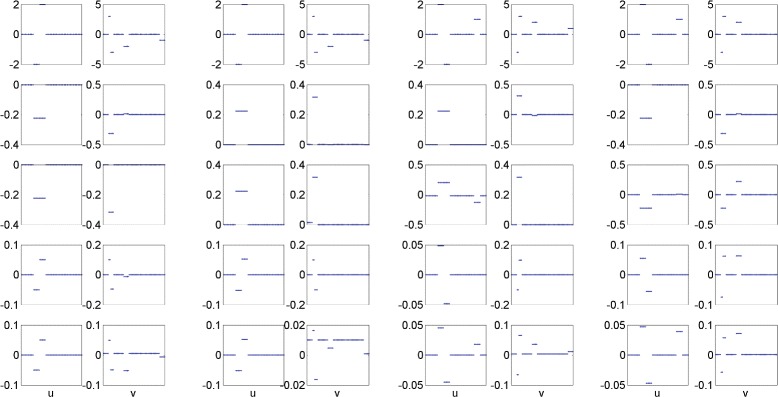
Canonical loadings estimated on four synthetic datasets. The first column is for Dataset 1, and the second column is for Dataset 2, and so forth. For each dataset, the weights of u are shown on the left panel, and those of v are on the right. The first row is the ground truth, and each remaining row corresponds to a specific method: (1) Ground Truth. (2) L1-SCCA. (3) FL-SCCA. (4) KG-SCCA. (5) GOSC-SCCA
Evaluation on real neuroimaging genetics data
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). For up-to-date information, see www.adni-info.org.
Table 2 contains the characteristics of the ADNI dataset used in this work. Participants including 568 non-Hispanic Caucasian subjects, including 196 healthy control (HC), 343 MCI and 28 AD participants. However, many participants’s data are incomplete due to various factors such as data loss. After cleaning those participants with incomplete information, we get 282 participants in our experiments. The genotype data were downloaded from LONI (adni.loni.usc.edu), and the preprocessed [11C] Florbetapir PET scans (i.e., amyloid imaging data) were also obtained from LONI. Before conducting the experiment, the amyloid imaging data had been preprocessed and the specific pipeline could be found in [14]. These imaging measures were adjusted by removing the effects of the baseline age, gender, education, and handedness via the regression weights derived from HC participants. We finally obtained 191 region-of-interest (ROI) level amyloid measurements which were extracted from the MarsBaR AAL atlas. We included four genetic markers, i.e., rs429358, rs439401, rs445925 and rs584007, from the known AD risk gene APOE. We intend to investigate if our GOSC-SCCA could identify this widely known associations between amyloid deposition and APOE SNPs.
Table 2.
Real data characteristics
| HC | MCI | AD | |
|---|---|---|---|
| Num | 196 | 343 | 28 |
| Gender(M/F) | 102/94 | 203/140 | 18/10 |
| Handedness(R/L) | 178/18 | 309/34 | 23/5 |
| Age (mean ±std.) | 74.77 ±5.39 | 71.92 ±7.47 | 75.23 ±10.66 |
| Education (mean ±std.) | 15.61 ±2.74 | 15.99 ±2.75 | 15.61 ±2.74 |
Shown in Table 3 are the 5-fold cross-validation results of various SCCA methods. We observe that GOSC-SCCA and KG-SCCA obtain similar correlation coefficients on every run, including the training performance and testing performance. Besides, they both are significantly better than L1-SCCA and FL-SCCA, which is consistent with the analysis in [14]. This result shows that GOSC-SCCA can improve the ability of identifying interesting imaging genetic associations compared with L1-SCCA and FL-SCCA.
Table 3.
5-fold cross-validation results on real data
| Methods | Training results | MEAN | Testing results | MEAN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1-SCCA | 0.50 | 0.50 | 0.53 | 0.53 | 0.54 | 0.52 | 0.56 | 0.61 | 0.45 | 0.47 | 0.38 | 0.49 |
| FL-SCCA | 0.44 | 0.43 | 0.46 | 0.45 | 0.46 | 0.45 | 0.49 | 0.56 | 0.39 | 0.43 | 0.37 | 0.45 |
| KG-SCCA | 0.53 | 0.52 | 0.55 | 0.54 | 0.56 | 0.54 | 0.56 | 0.61 | 0.47 | 0.52 | 0.45 | 0.52 |
| GOSC-SCCA | 0.53 | 0.52 | 0.55 | 0.55 | 0.56 | 0.54 | 0.56 | 0.62 | 0.47 | 0.51 | 0.45 | 0.52 |
The estimated correlation coefficients and their MEAN are shown. The best correlation coefficients and those that are NOT significantly worse than the best ones (t-test with p-value smaller than 0.05) are shown in bold
Figure 2 contains the estimated canonical loadings obtained from 5-fold cross-validation. To facilitate the interpretation, we employ the heat map for this real data. Each row denotes a method, and u (genetic markers) is shown on the left panel and v (imaging markers) is on the right. As we can see, on the genetic side, all four SCCA exhibit similar canonical loading pattern. Since every SCCA here incorporates the lasso (ℓ1-norm), they select only the APOE e4 SNP (rs429358), which is a widely known AD risk marker, with those irrelevant ones discarded to assure sparsity. On the imaging side, L1-SCCA identifies many signals which is hard to interpret. FL-SCCA fuses those adjacent features together due to its pairwise smoothness, which can be easily observed from the figure. But it is difficult to interpret either. GOSC-SCCA and KG-SCCA perform similarly again in this run. They both identify the imaging signals in accordance with the findings in [20]. It is easily to observe that they estimated a very clean signal pattern, and thus is easy to conduct further investigation. Recall the results in Table 3, the association between the marker rs429358 and the amyloid accumulation in the brain is relatively strong, and thus the signal can be well captured by both KG-SCCA and GOSC-SCCA. In addition, the correlations among the imaging variables and those among genetic variables are high enough so that the signs of these correlations can hardly be impeded by the noises. That is, the signs of sample correlations tend to be correctly estimated. Therefore, KG-SCCA does not suffer sign directionality issue, and so performs similarly to GOSC-SCCA. However, if some sample correlations are not very strong and their signs are mis-estimated, KG-SCCA may not work very well (see the results of the second synthetic dataset). In summary, this reveals that our method has better generalization ability, and could identify biologically meaningful imaging genetic associations.
Fig. 2.
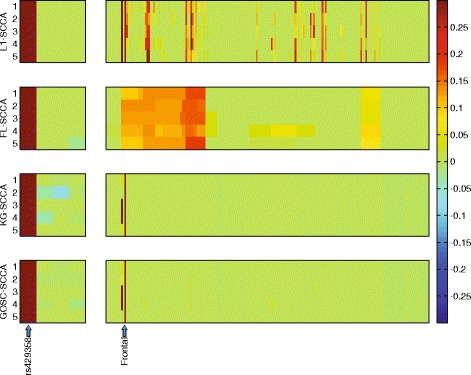
Canonical loadings estimated on the real dataset. Each row corresponds to a SCCA method: (1) L1-SCCA. (2) FL-SCCA. (3) KG-SCCA. (4) GOSC-SCCA. For each row, the estimated weights of u are shown on the left figure, and those of v on the right
Discussion
In this paper, we have proposed a structured SCCA method GOSC-SCCA, which intended to reduce the estimation bias caused by the incorrect sign of sample correlation. GOSC-SCCA employed the GOSCAR (Graph OSCAR) regularizer which is an extension of the popular penalty OSCAR. The GOSC-SCCA could pull those highly correlated features together no matter that they were positively correlated or negatively correlated. We also provide a theoretical quantitative description of the grouping effect of our SCCA method. An effective algorithm was also proposed to solve the GOSC-SCCA problem and the algorithm was guaranteed to converge.
We evaluated GOSC-SCCA and three other popular SCCA methods on both synthetic datasets and a real imaging genetics dataset. The synthetic datasets consisted of different ground truth, i.e. different correlation coefficients and canonical loadings. GOSC-SCCA was capable of consistently identifying strong correlation coefficients on both training set and testing set, and either outperformed or performed similarly to the competing methods. Besides, GOSC-SCCA successfully and accurately recognized the signals which were the closest to the ground truth when compared with the competing methods.
The results on the real data showed that both GOSC-SCCA and KG-SCCA could find an important association between the APOE SNPs and the amyloid burden measure in the frontal region of the brain. KG-SCCA performs similarly to GOSC-SCCA on this real data largely because of the strong correlations between the variables within the genetic data, as well as those within the imaging data. In this case, the signs of the correlation coefficients between these variables tend to be correctly calculated, and so KG-SCCA does not have the sign directionality issue. On the other hand, if the correlations among some variables are not very strong, the performance of KG-SCCA can be affected by the mis-estimation of some correlation signs. In this case, GOSC-SCCA, which is designed to overcome the sign directionality issue, is expected to perform better than KG-SCCA. This fact has already been validated by the results of the second synthetic dataset.
The satisfactory performance of GOSC-SCCA, coupled with its theoretical convergence and grouping effect, demonstrates the promise of our method as an effective structured SCCA method in identifying meaningful bi-multivariate imaging genetic associations. The following are a few possible future directions. (1) Note that the identified pattern between the APOE genotype and amyloid deposition is a well-known and relatively strong imaging genetic association. Thus one direction is to apply GOSC-SCCA to more complex imaging genetic data for revealing novel but less obvious associations. (2) The data tested in this study is brain wide but targeted only at APOE SNPs. Another direction is to apply GOSC-SCCA to imaging genetic data with higher dimensionality, where more effective and efficient strategies for parameter tuning and cross-validation warrant further investigation. (3) The third direction is to employ GOSC-SCCA as a knowledge-driven approach, where pathways, networks or other relevant biological knowledge can be incorporated in the model to aid association discovery. In this case, comparative study can also been done between GOSC-SCCA and other state-of-the-arts knowledge-guided SCCA methods in bi-multivariate imaging genetics analyses.
Conclusions
We have presented a new structured sparse canonical analysis (SCCA) model for analyzing brain imaging genetics data and identifying interesting imaging genetic associations. This SCCA model employs a regularization item based on the graph octagonal selection and clustering algorithm for regression (GOSCAR). The goal is twofold: (1) encourage highly correlated features to have similar canonical weights, and (2) reduce the estimation bias via removing the requirement of pre-defining the sign of the sample correlation. As a result, it could pull highly correlated features together no matter whether they are positively or negatively correlated. Empirical results on both synthetic and real data have demonstrated the promise of the proposed method.
Acknowledgements
At Indiana University, this work was supported by NIH R01 LM011360, U01 AG024904, RC2 AG036535, R01 AG19771, P30 AG10133, UL1 TR001108, R01 AG 042437, R01 AG046171, and R03 AG050856; NSF IIS-1117335; DOD W81XWH-14-2-0151, W81XWH-13-1-0259, and W81XWH-12-2-0012; NCAA 14132004; and CTSI SPARC Program. At University of Texas at Arlington, this work was supported by NSF CCF-0830780, CCF-0917274, DMS-0915228, and IIS-1117965. At University of Pennsylvania, the work was supported by NIH R01 LM011360, R01 LM009012, and R01 LM010098.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neu-roimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengi-neering, and through generous contributions from the following: AbbVie, Alzheimer’s Asso-ciation; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Bio-gen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Develop-ment LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Neu-roRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeu-tics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the Na-tional Institutes of Health (www.fnih.org). The grantee organization is the Northern Califor-nia Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are dis-seminated by the Laboratory for Neuro Imaging at the University of Southern California.
Declarations
Publication charges for this article have been funded by the corresponding author.
This article has been published as part of BMC Systems Biology Volume 10 Supplement 3, 2016: Selected articles from the International Conference on Intelligent Biology and Medicine (ICIBM) 2015: systems biology. The full contents of the supplement are available online at
http://bmcsystbiol.biomedcentral.com/articles/supplements/volume-10-supplement-3.
Availability of data and materials
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu).
Authors contributions
LD, JM, AS, and LS: overall design. LD, HH, and MI: modeling and algorithm design. LD and JY: experiments. SK, SR, and AS: data preparation and result evaluation. LD, JY and LS: manuscript writing. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Footnotes
Alzheimer’s Disease Neuroimaging Initiative, Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Contributor Information
Lei Du, Email: leidu@iu.edu.
Heng Huang, Email: heng@uta.edu.
Jingwen Yan, Email: jingyan@umail.iu.edu.
Sungeun Kim, Email: sk31@iupui.edu.
Shannon Risacher, Email: srisache@iupui.edu.
Mark Inlow, Email: inlow@rose-hulman.edu.
Jason Moore, Email: jhmoore@exchange.upenn.edu.
Andrew Saykin, Email: asaykin@iupui.edu.
Li Shen, Email: shenli@iu.edu.
References
- 1.Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. NeuroImage. 2010;53(3):1147–59. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkhomenko E, Tritchler D, Beyene J. Sparse canonical correlation analysis with application to genomic data integration. Stat Appl Genet Mol Biol. 2009;8(1):1–34. doi: 10.2202/1544-6115.1406. [DOI] [PubMed] [Google Scholar]
- 3.Witten DM, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10(3):515–34. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Liu H, Carbonell JG. Structured sparse canonical correlation analysis. In: International Conference on Artificial Intelligence and Statistics, JMLR Proceedings 22, JMLR.org: 2012.
- 5.Chen X, Liu H. An efficient optimization algorithm for structured sparse cca, with applications to eqtl mapping. Stat Biosci. 2012;4(1):3–26. doi: 10.1007/s12561-011-9048-z. [DOI] [Google Scholar]
- 6.Chi EC, Allen G, Zhou H, Kohannim O, Lange K, Thompson PM, et al. Imaging genetics via sparse canonical correlation analysis. In: Biomedical Imaging (ISBI), 2013 IEEE 10th Int Sym On: 2013. p. 740–3, doi:http://dx.doi.org/10.1109/ISBI.2013.6556581. [DOI] [PMC free article] [PubMed]
- 7.Lin D, Calhoun VD, Wang YP. Correspondence between fMRI and SNP data by group sparse canonical correlation analysis. Medical image analysis. 2014;18(6):891–902. doi: 10.1016/j.media.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L, Yan J, Kim S, Risacher SL, Huang H, Inlow M, Moore JH, Saykin AJ, Shen L. International Conference on Medical Image Computing and Computer Assisted Intervention. Berlin, Germany: Springer; 2014. A novel structure-aware sparse learning algorithm for brain imaging genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8(1):1–27. doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Bushman FD, et al. Structure-constrained sparse canonical correlation analysis with an application to microbiome data analysis. Biostatistics. 2013;14(2):244–58. doi: 10.1093/biostatistics/kxs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bondell HD, Reich BJ. Simultaneous regression shrinkage, variable selection, and supervised clustering of predictors with oscar. Biometrics. 2008;64(1):115–23. doi: 10.1111/j.1541-0420.2007.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24(9):1175–82. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- 13.Yang S, Yuan L, Lai YC, Shen X, Wonka P, Ye J. Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, USA: ACM; 2012. Feature grouping and selection over an undirected graph. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan J, Du L, Kim S, Risacher SL, Huang H, Moore JH, Saykin AJ, Shen L. Transcriptome-guided amyloid imaging genetic analysis via a novel structured sparse learning algorithm. Bioinformatics. 2014;30(17):564–71. doi: 10.1093/bioinformatics/btu465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardoon D, Szedmak S, Shawe-Taylor J. Canonical correlation analysis: An overview with application to learning methods. Neural Comput. 2004;16(12):2639–64. doi: 10.1162/0899766042321814. [DOI] [PubMed] [Google Scholar]
- 16.Nie F, Huang H, Cai X, Ding CH. Advances in Neural Information Processing Systems. Massachusetts, USA: The MIT Press; 2010. Efficient and robust feature selection via joint 2, 1-norms minimization. [Google Scholar]
- 17.Grosenick L, Klingenberg B, Katovich K, Knutson B, Taylor JE. Interpretable whole-brain prediction analysis with graphnet. NeuroImage. 2013;72:304–21. doi: 10.1016/j.neuroimage.2012.12.062. [DOI] [PubMed] [Google Scholar]
- 18.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Royal Stat Soc Ser B (Stat Method) 2005;67(2):301–20. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 19.Lorbert A, Eis D, Kostina V, Blei DM, Ramadge PJ. Exploiting covariate similarity in sparse regression via the pairwise elastic net. In: International Conference on Artificial Intelligence and Statistics, JMLR Proceedings 9, JMLR.org: 2010. p. 477–84.
- 20.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, Foroud TM, Hakonarson H, Huentelman MJ, Aisen PS, et al. Apoe and bche as modulators of cerebral amyloid deposition: a florbetapir pet genome-wide association study. Mole psychiatry. 2014;19(3):351–7. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu).


