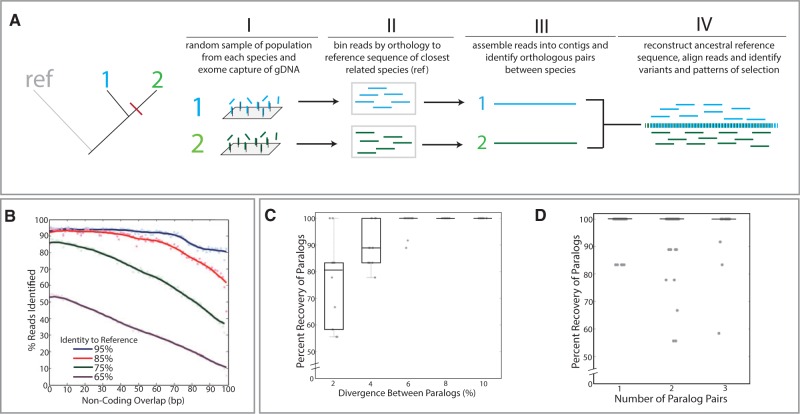
Fig. 1.
Phylomapping approach for analysis of character change in species lacking prior genetic resources. (A) Overview of approach. In this example, there are two species, “1” and “2,” with a common outgroup, “ref,” that has existing genetic resources from which to design the targeted capture array (I) and guide identification of reads (II). Reads are binned by orthology and assembled into contigs (III). Pairwise analysis between orthologous contigs enables reconstruction of a shared ancestral sequence that is then used to align and compare sequence variance (IV). (B–D) Test limits of approach through simulated assembly and annotation of reads from divergent genomes. (B) In silico 100-bp reads were generated at varying levels of divergence from the zebrafish reference genome and put into the phylomapping pipeline, utilizing the zebrafish reference genome to scaffold the analysis. Alignment and identification of reads from varying levels of divergence relative to the reference genome showing read recovery with high levels of variation and extending into noncoding sequence flanking the exons. Lines represent kernel regression. Simulated reconstruction of paralogs from mixed read data (supplementary fig. S1, Supplementary Material online) as a function of the divergence between paralogous sequences (C) and the number of paralogs present within the mixed read pools (D). Each dot represents an individual simulation.