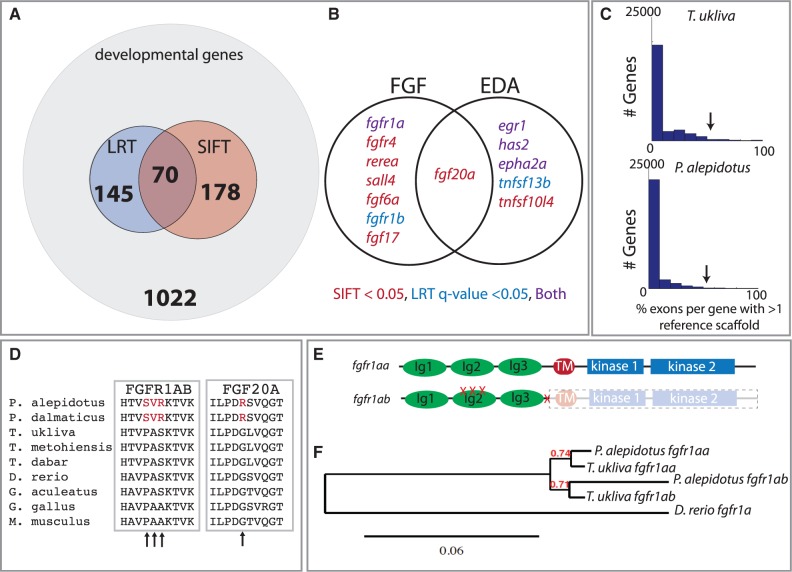
Fig. 3.
Selection on an FGF ligand-receptor pair within Phoxinellus underlying scale reduction. (A) Analysis of a subset of developmental genes as defined by select gene ontology terms (supplementary table S1, Supplementary Material online) with exons showing signatures of nonsense or deleterious SNPs (SIFT) and accelerated sequence evolution relative to neutral drift (LRT) (supplementary table S1, Supplementary Material online). (B) Genes within the EDA and FGF signaling pathways showing either LRT q value < 0.05 or SIFT < 0.05 (supplementary table S1, Supplementary Material online). (C) Histogram of the percentage of exons per gene showing a signature of duplication (supplementary table S3, Supplementary Material online); arrow indicates location of fgfr1a. (D) Multiple protein alignment of Fgfr1ab and Fgf20a. Arrows indicate amino acid changes within the Phoxinellus genus. (E) Location of Phoxinellus fgfr1ab nonsynonymous mutations and premature truncation mapped onto Fgfr1a protein (supplementary fig. S2, Supplementary Material online). (F) Reconstructed gene tree from cDNA of fgfr1aa and fgfr1ab transcripts isolated from Phoxinellus alepidotus skin and Telestes ukliva exome sequencing data.