Fig. 1.
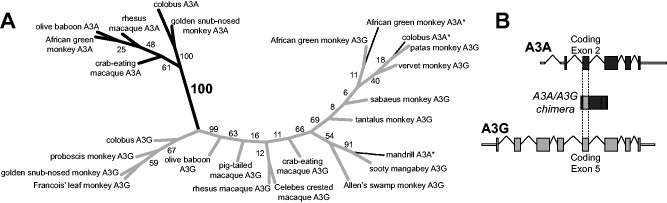
Primate A3A-A3G chimeras contain exon 5 of A3G in place of A3A exon 2. (A) We generated an unrooted maximum likelihood phylogeny using an alignment of A3A exon 2, and A3G exon 5 nucleotide sequences (see Materials and Methods for details). Branches leading to A3A, A3G, or chimeric sequences are shown in black, gray, and dotted lines, respectively. Bootstrap values represent percentages based on 1,000 iterations. The previously reported “alternative exon 2-containing” A3As from mandrill, AGM, colobus (marked *) group with A3Gs, whereas the newly sequenced, corrected A3A exon 2s group with the rest of the A3As. Although the phylogeny is not well resolved within the A3A and A3G clades, the bootstrap support for separation of these two clades is high, grouping the chimeric A3A* sequences with the A3Gs. (B) The chimeric A3A-A3Gs comprise A3G coding exon 5 swapped into A3A in place of the second coding exon of A3A. These chimeras resulted from sequence similarity between the introns on either side of these two exons.