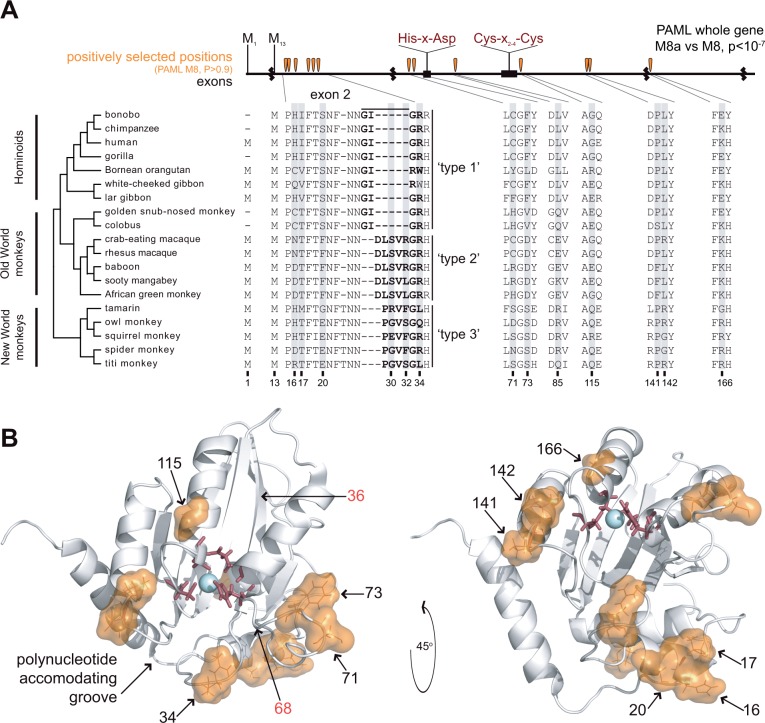
Fig. 2.
Sequence variation and positive selection in primate A3A. (A) We created an alignment of 19 A3A sequences spanning the hominoids, Old World monkeys, and New World monkeys using publicly available databases and sequencing. Several primates have lost the methionine at “position 1” and instead initiate translation using the methionine at “position 13”. Exon boundaries are denoted by black squiggly lines and the conserved deaminase motif is noted in red labels, top. The exon 2 sequence of A3A varies among primates, which we classify into three general “types.” We tested the signature of positive selection using PAML’s NSsites maximum likelihood method and observe a significant signature of gene-wide positive selection (PAML M8a vs. M8). We also identify 13 codons with statistically significant positive selection (PAML M8, BEB, P > 0.9, table 1), as indicated by shading in the protein alignment and orange teardrops in the exon diagram. (B) The positively selected positions are shown as orange space-filling spheres with positions numbered in black on the white ribbon diagram (A3A NMR structure, PDB ID 2M65 [Byeon et al. 2013]); active site residues are shown in red sticks. Positively selected positions cluster in secondary structure as well tertiary structure along the “polynucleotide accommodating groove” (Bulliard et al. 2011). Amino acid positions numbered in red were shown to increase restriction of LINE-1 by human A3G when swapped from A3A into A3G (Bulliard et al. 2011).