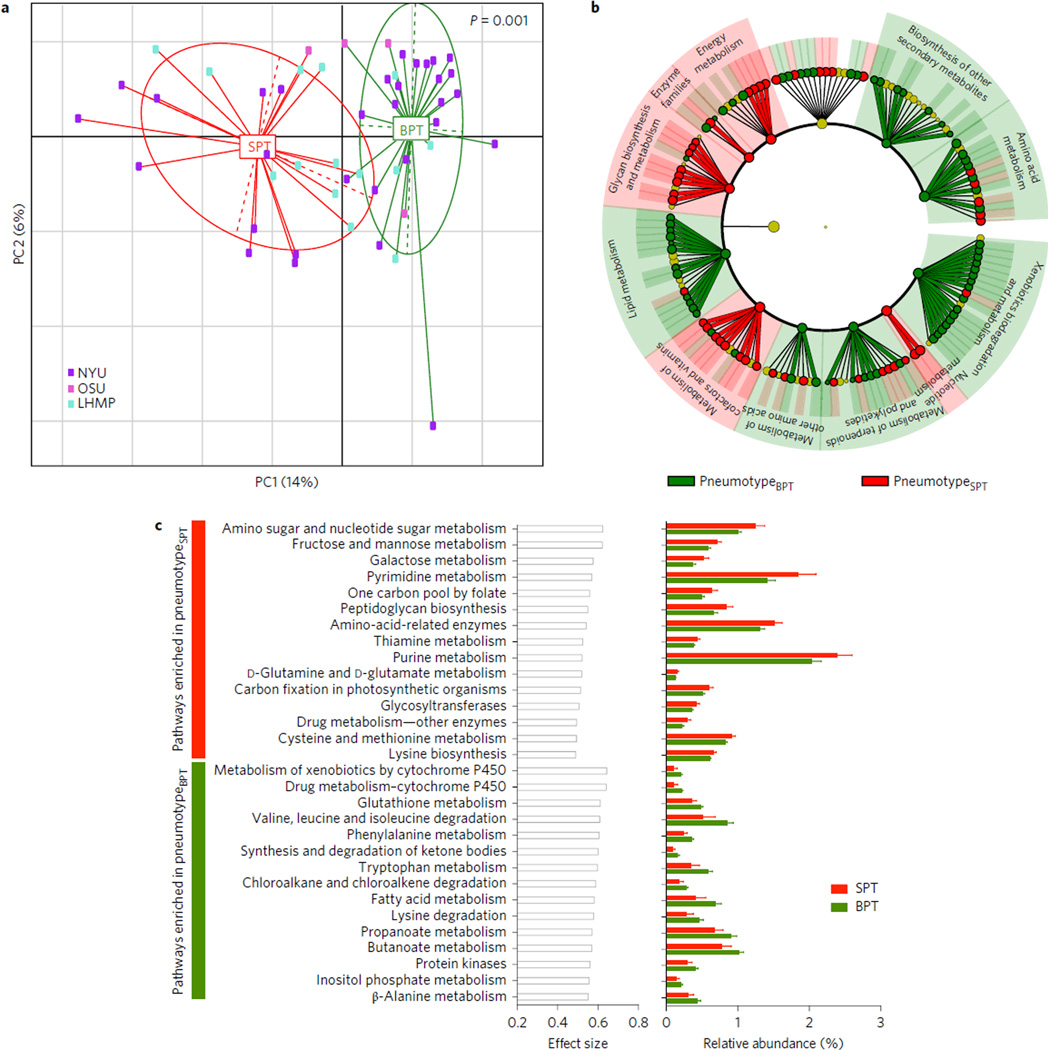
Figure 2. Comparison of inferred metagenomes of pneumotypeSPT and pneumotypeBPT.
a, PCoA based on Jensen–Shannon divergence shows that the metagenome of pneumotypeSPT is significantly different from the metagenome of pneumotypeBPT. b, LEfSe analysis was performed using the summarized functional annotation for the KOs annotated to metabolism inferred for each BAL sample. This analysis showed multiple functional differences in the genomic composition of pneumotypeSPT as compared with pneumotypeBPT. c, STAMP was used to determine metabolic pathways differentially enriched (P < 0.05) and their effect size (η2). The 15 top metabolic pathways for each pneumotype are represented with effect size and relative abundance.