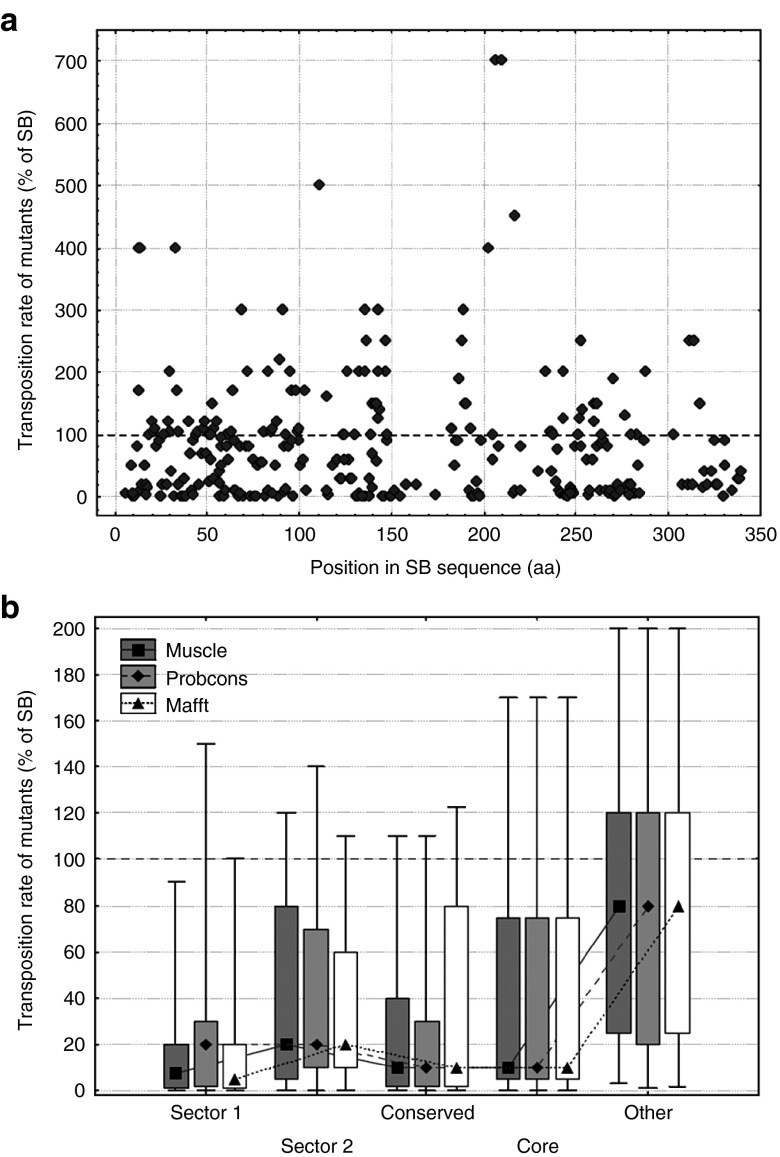
Figure 2.
Effect of residue location on the transposition rate of SB mutants. (a) The location of the mutations along the SB transposase sequence, and their effect on transposition rate. The 286 mutants are distributed approximately evenly across the sequence; the majority of mutants reduces transposition rate (< 100% of SB). None of the Pfam conserved domains show a clear difference from the rest of the sequence. (b) The effect of sectors, conserved residues and protein core on transposition rate (median, box: 25–75%, whiskers: 10–90%). Mutants in both sectors, conserved residues and residues of the protein core have significantly lower transposition rates than other residues, irrespectively of the aligner used (P < 0.05, Mann-Whitney U-tests).