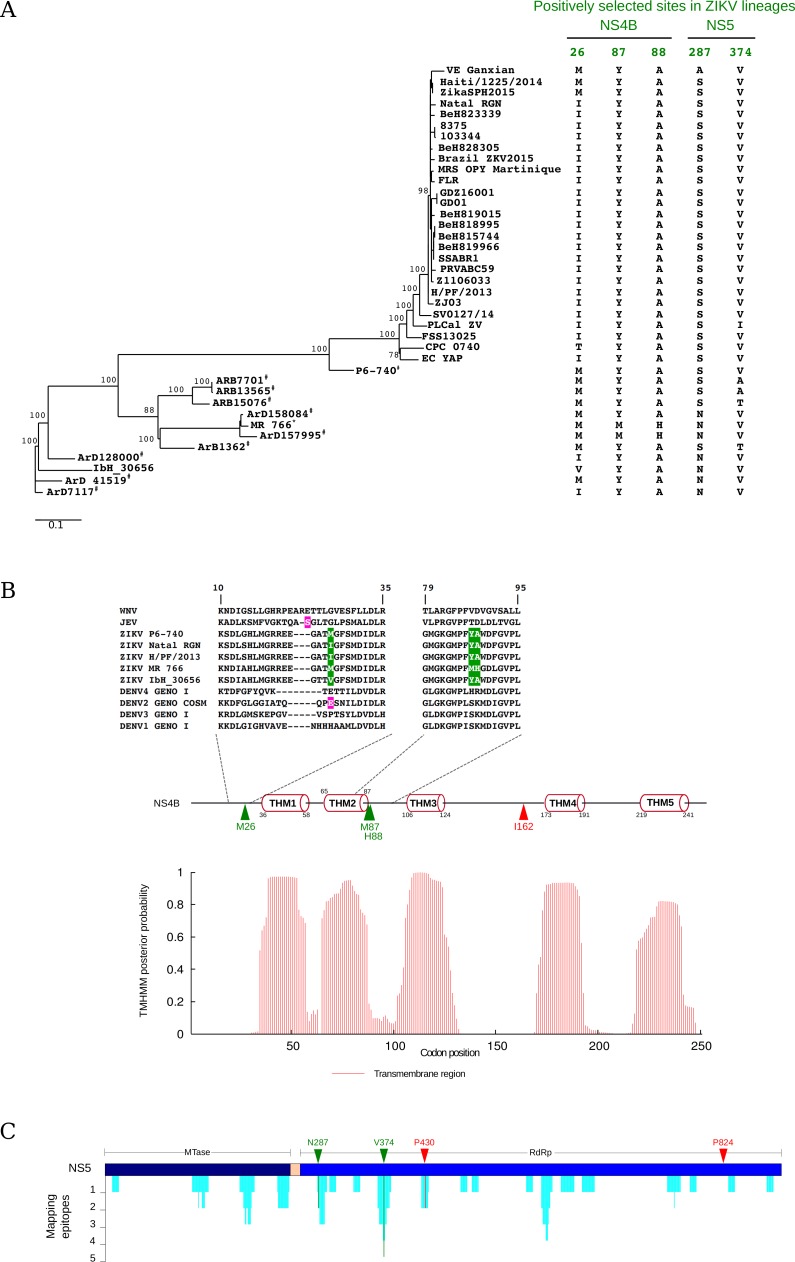
Fig 3. Ongoing positive selection in ZIKV isolates.
(A) Maximum likelihood phylogeny for the nonstructural region (nucleotides 2371 to 8994). Amino acids at the five selected sites are shown for all ZIKV sequences. Viruses isolated from mosquitos and macaques are denoted with hash and asterisk symbols, respectively. Branch length is proportional to nucleotide substitutions per codon. Bootstrap values for internal branches >75% are shown. The phylogenetic tree is unrooted. (B) TMHMM prediction of transmembrane helices (TMH1-5) for the ZIKV NS4B protein and schematic representation of protein topology. Positively selected sites in the flavivirus phylogeny and in ZIKV strains are indicated by red and green triangles, respectively. Amino acid alignments of the regions surrounding selected sites are shown for 5 representative ZIKV, for DENV sequences belonging to the four serotypes, for JEV (NC_001437), and for WNV (strain NY-99, NC_001563). In the alignment, positively selected sites in ZIKV are shown in green; sites that are positively selected in other flaviviruses are marked in magenta. (C) Immune epitope mapping on NS5. Cyan bars indicate the number of epitopes overlapping each NS5 residue. Positively selected sites are colored as above. Mtase: methyltransferase domain; RdRp: RNA-dependent RNA polymerase domain.