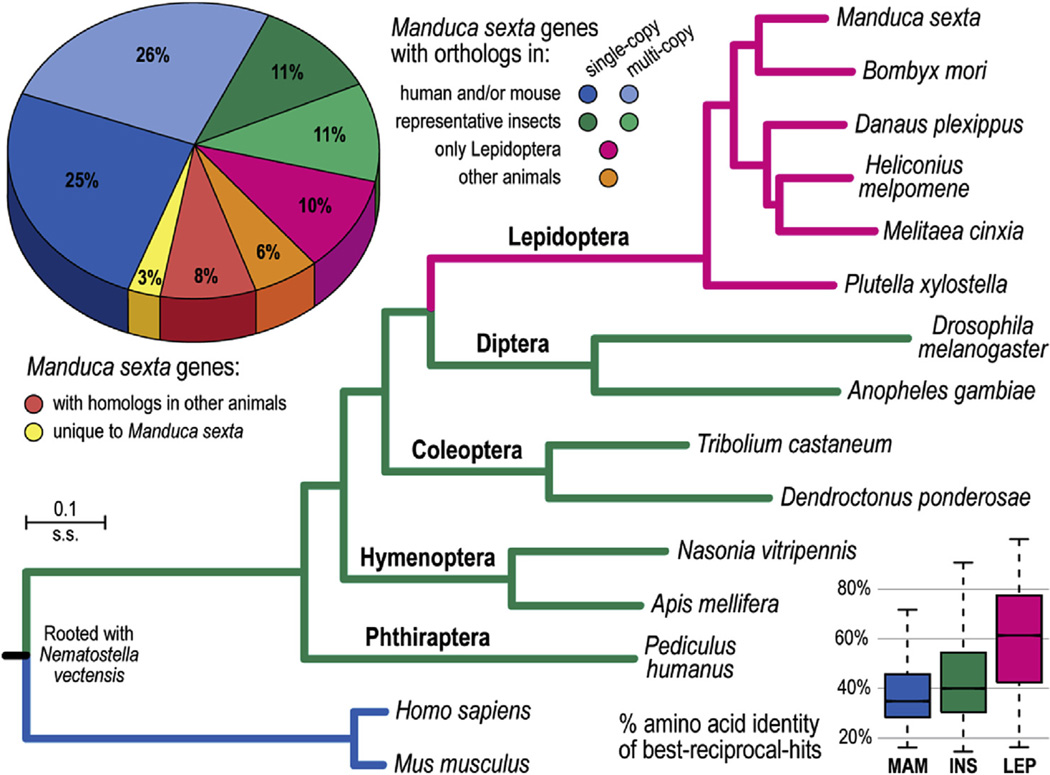
Fig. 1.
The Manduca sexta gene repertoire and molecular species phylogeny. Approximately half of the 15,451 M. sexta genes have identifiable orthologs in the representative genomes of mammals, human and mouse (pie chart, blue), suggesting that these are ancient genes likely to have been present in the metazoan ancestor. A further 32% of M. sexta genes exhibit orthology to genes from the other seven representative insect species (green, 22% or 3427 genes), or only to genes from the other five lepidopteran species (pink, 10% or 1588 genes). Of the remaining genes, some have orthologs (orange) or homologs (red) in other animal species (other metazoan species from OrthoDB), leaving 417 M. sexta genes (app. 3%) without any recognizable homologs (yellow, e-value cutoff 1e-3). Employing aligned protein sequences of universal single-copy orthologs to estimate the molecular species phylogeny rooted with the starlet sea anemone, Nematostella vectensis, shows that the Lepidoptera and Diptera exhibit the fastest rates of molecular divergence. All nodes have 100% bootstrap support. The boxplots show the distributions of percent amino acid identities between M. sexta proteins and their best-reciprocal-hits from mammal species (MAM; median 34.9%), insect species (INS; median 40.1%), and lepidopteran species (LEP; median 60.2%). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)