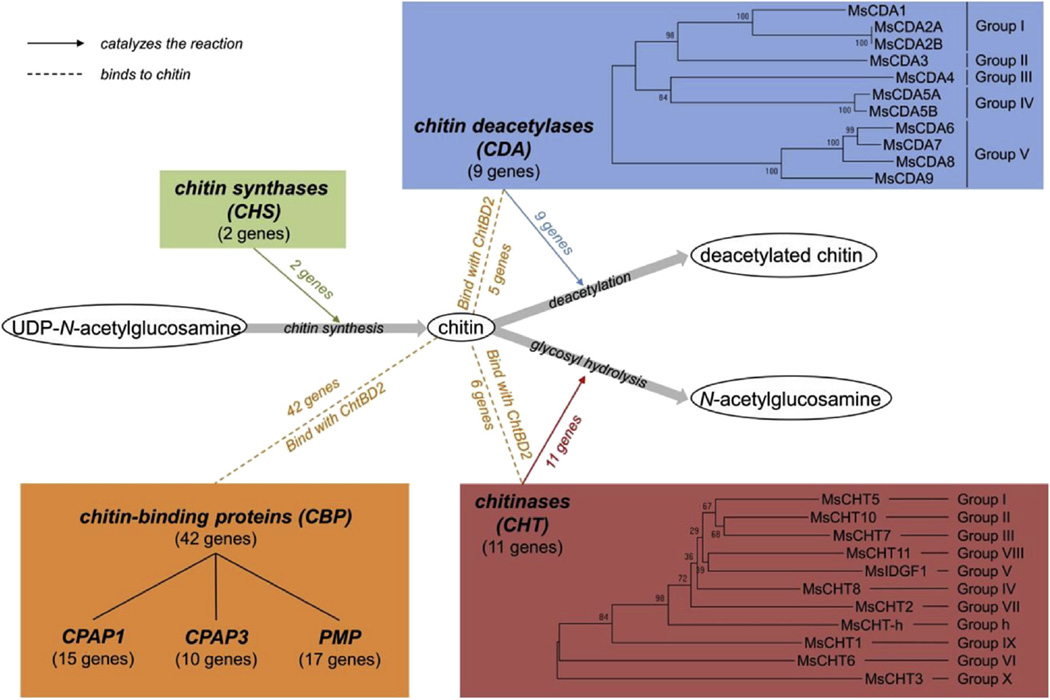
Fig. 6.
Summary of the families of genes coding for chitin metabolism enzymes and chitin binding proteins (CBPs) in the M. sexta genome. Dotted lines indicate the binding of CBPs to chitin by their chitin binding domains (CBD – orange), and lines with an arrow indicate that chitin metabolism enzymes and their functions in chitin synthesis (chitin synthases, CHS – green), deacetylation (chitin deacetylases, CDA – blue) and degradation (chitinases, CHT – red). For CDA and CHT, a phylogenetic tree has been constructed using the neighbor-joining method, with 1000 replications of bootstrap analyses, implemented in MEGA 6.06 (Tamura et al., 2011). CPAP: cuticular proteins analogous to peritrophin; PMP: peritrophic matrix protein. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)