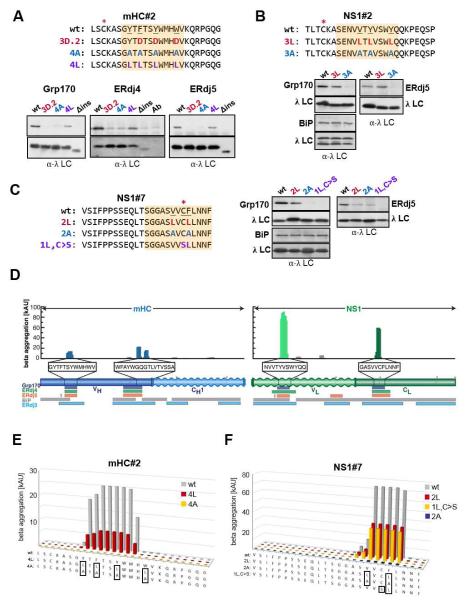
Figure 4. Grp170, ERdj4 and ERdj5 binding sites correspond to aggregation-prone regions in their clients.
(A-C) Cells expressing the indicated (co-)chaperones together with mHC#2 (A), NS1#2 (B) or NS1#7 (C) peptide constructs were analyzed by immunoprecipitation-coupled western blotting as described in Figure 3F. The 12-aa fragments identified in Figure 3 and S3 are indicated with orange boxes. The top panel in each set shows anti-(co-)chaperone western blots and the bottom anti-λ blots. For ERdj4, a separate lane with the anti-λ antibody was included and samples were run under-non-reducing conditions to reduce background signal from the precipitating antibody. (D) Sequences for the mini-HC (blue) and NS1 LC (green) were subjected to analysis by the TANGO algorithm, which identifies regions that are predicted to be prone to β-aggregate formation. Wavy segments indicate unfolded domains in each client (when expressed in isolation). The core binding peptides for Grp170 (purple), ERdj4 (green), and ERdj5 (orange) are indicated below, as are binding sites for BiP (grey) and ERdj3 (blue). (E-F) The TANGO algorithm was used to calculate the β-aggregation potential for the indicated mHC#2 (E) and NS1#7 (F) constructs. Each mutant is color-coded with sequences indicated below and mutated residues boxed.