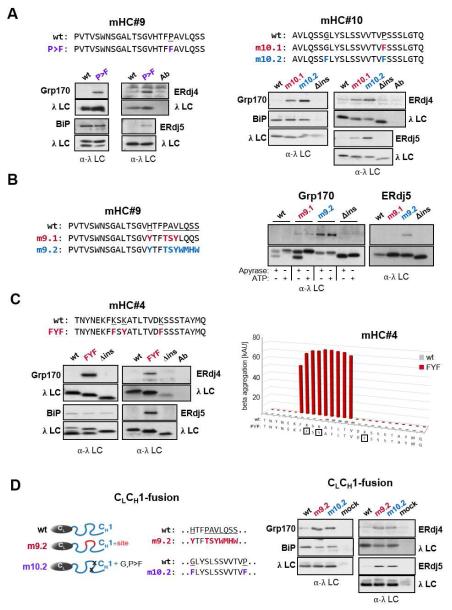
Figure 5. Mutations predicted to increase aggregation lead to (co-)chaperone binding and can result in aggregation of full-length clients.
(A) Putative (co-)chaperone binding sites were introduced into mHC#9 and mHC#10 with the indicated substitutions and analyzed by immunoprecipitation-coupled western blotting for (co-)chaperone binding as before. (B) The indicated substitutions were made in mHC#9 to create a partial (m9.1) or the complete (m9.2) binding site identified in mHC#2. Constructs were analyzed as in (A). In the case of Grp170 binding samples were lysed with apyrase or ATP added. (C) The TANGO algorithm was used to predict amino acid changes that would lead to an aggregation-prone region in mHC#4, which was negative for both BiP and (co-)chaperone binding. The wild-type and mutant constructs were examained for BiP and (co-)chaperone binding as above. (D) Mutations that led to (co-)chaperone binding in either mHC#9 or #10 were engineered into the corresponding region of the CH1 domain in a chimeric protein composed of the ER targeted λ CL domain and the γ1 CH1 domain. (Co-)chaperone interactions and BiP binding were detected by immunoprecipitation-coupled western blotting.