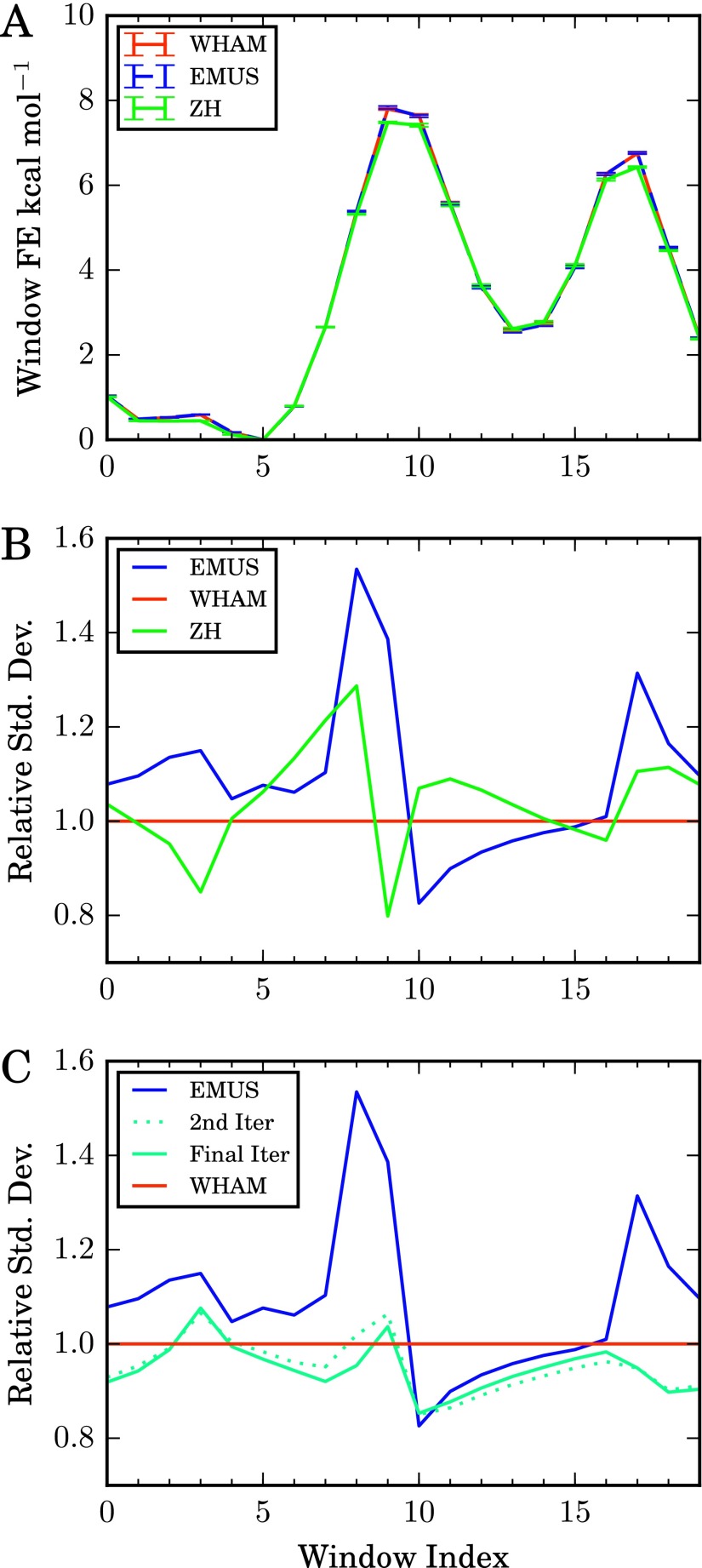
FIG. 1.
Comparison of umbrella sampling methods applied to simulation data for the alanine dipeptide. (a) Average window free energies, Gi, for the indicated methods. Error bars are estimated standard deviations of the means. (b) Standard deviation of each method relative to that of the WHAM algorithm. Colors are the same as in (a). (c) EMUS as the first step in a self-consistent iteration to solve the MBAR equations (see text). The number of uncorrelated samples in each window (ni) was estimated by calculating the integrated autocorrelation of the ϕ dihedral angle from each trajectory. Results shown are for identical molecular dynamics data (see text for simulation details); the methods differ only with respect to combination of the data to estimate the free energies.