FIG 2.
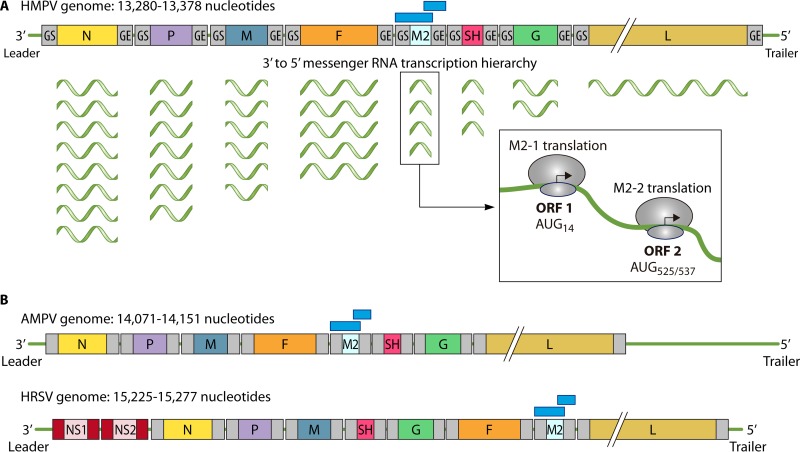
(A) Schematic structure of the HMPV ssRNA genome and arrangement of each of the eight genes within the genome. Each gene is flanked by two conserved sequences, which are referred to as the gene start (GS) and gene end (GE) sequences. During transcription, the TR complex stops at each GE to restart transcription in the following GS sequence (84). However, because the TR complex may stall, this process leads to an increased abundance of mRNAs closer to the 3′ leader. At the bottom right is a sketch of the translation mechanism producing two polypeptides from the M2 mRNA. Two different ORFs located at position 14 and position 525/537 are used to synthesize the M2-1 and M2-2 proteins, respectively (85). (B) Schematic compositions of the AMPV and HRSV genomes. Note the striking degree of conservation of gene order between HMPV and AMPV. Sequence analyses indicate that while most AMPV and HMPV structural proteins display ∼80% amino acid identity, the G and SH proteins display high amino acid variability ranging from 70 to 80% (78, 264). Also, note that the genomes of HMPV and AMPV lack homologous genes for the HRSV NS1 and NS2 proteins (highlighted in red). In panels A and B, the genomes are not illustrated to scale.