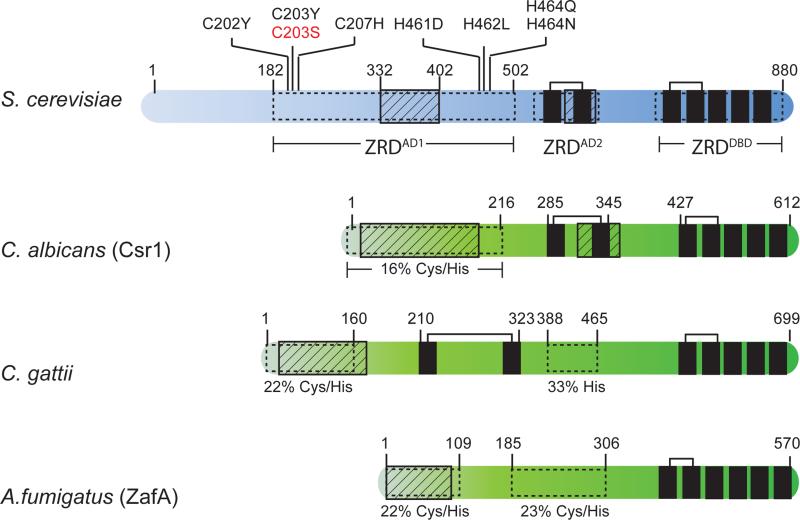
Figure 1.
Domain structures of Zap1 from S. cerevisiae, C. albicans, C. gattii, and A. fumigatus. Zap1 homologs in C. albicans and A. fumigatus are named Csr1 and ZafA respectively. Features highlighted in Zap1 from S. cerevisiae include the acidic rich activation domains (striped rectangles), zinc-responsive domains (white rectangles), and zinc finger domains (black rectangles). tCWCH2 zinc fingers are connected by brackets. Amino acid substitutions that disrupt the zinc-dependent inactivation of ZRDAD1 are also indicated. The original Zap1-1up allele is shown in red. Additional features highlighted in other Zap1 homologs include regions rich in acidic residues (striped rectangles) and cysteine and histidine residues (white rectangles). Numbers above each protein represent amino acid position.