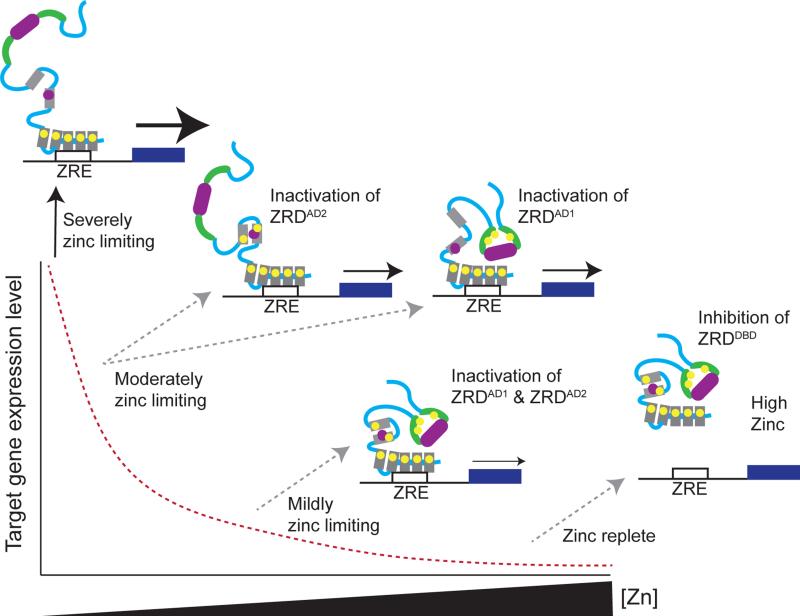
Figure 2.
Predicted model of Zap1 regulation in S. cerevisiae. Under severely zinc-limiting conditions, Zap1 target gene expression is maximally induced. Under these conditions Zap1 is bound to ZREs in target gene promoters, and AD1 and AD2 recruit coactivator complexes to drive target gene expression. Under moderately zinc-limiting conditions, the ZF1/ZF2 tCWCH2 zinc fingers bind zinc leading to the inactivation of AD2, or the cysteine/histidine rich region of ZRDAD1 binds zinc leading to the inactivation of AD1. Both of these events lead to reduced target gene expression. Under mildly zinc-limiting conditions, inactivation of ZRDAD1 and ZRDAD2 further reduces Zap1 target gene expression. When zinc is in excess, Zap1 dissociates from DNA, preventing Zap1-dependent transcription. Features shown include zinc finger domains (gray boxes), zinc ions (yellow circles), activation domains (purple rectangles with +'s), and the cysteine/histidine rich regions of AD1 (green crescents). The red dashed line represents Zap1 target gene expression in response to zinc.