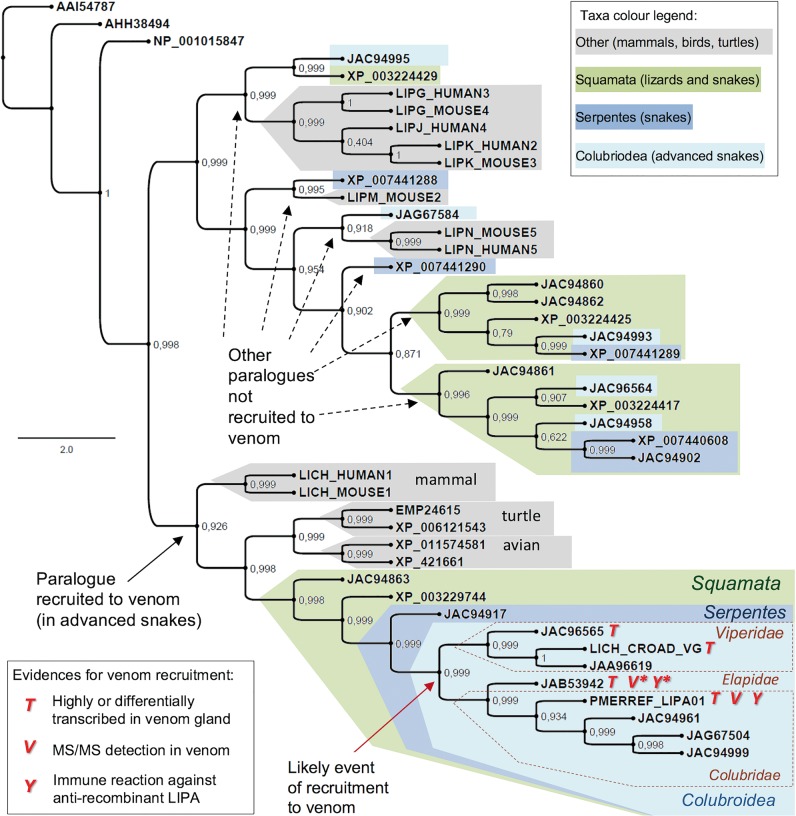
Fig. 7.—
Bayesian tree of acid lipase proteins from vertebrates focusing on the existence of various paralogues in squamata reptiles with a possible recruitment of one paralogue to venom in advanced snakes. The protein sequences are referred by their accession numbers. *JAB53942 is from M. fulvius but the proteomic evidence was obtained from similar sequence from M. altirostris (Correa-Netto et al. 2011) and the immunochemical evidence is from M. corallinus venom (this work). Numbers on the nodes represent Bayesian posterior probabilities. Bayesian inferences was obtained by MrBayes software version 3.1.1 30, utilizing 1 × 106 numbers of generations for the Markov chain Monte Carlo algorithm in four chains, sampled every 100 and using the aamodelpr prior set to mixed models.