Fig. 1.—
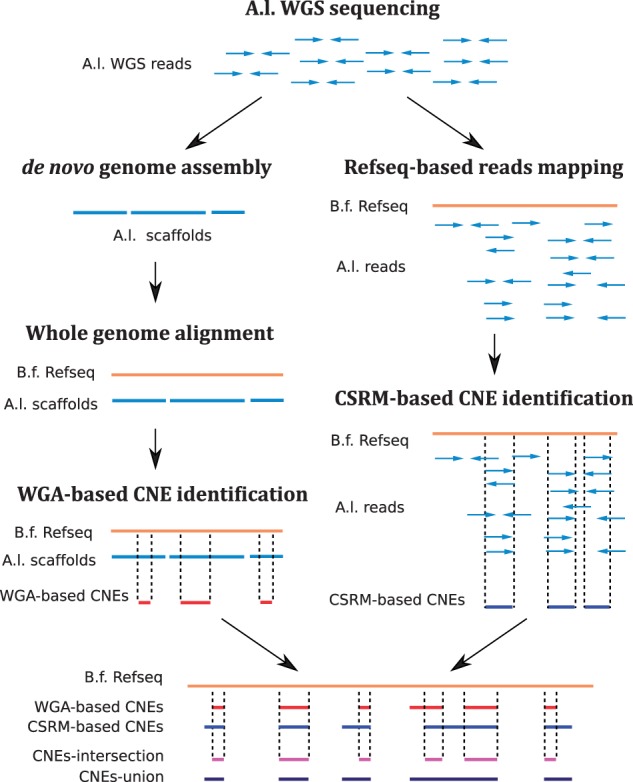
A diagram to show the two parallel strategies used in this study for CNE identification. Starting from the raw reads, a whole genome assembly of A. lucayanum was generated and further aligned to the B. floridae reference genome for CNE identification. We refer CNEs identified by this way as whole-genome alignment-based CNEs (WGA-based CNEs). Alternatively, another CNE set was generated by directly mapping the A. lucyanuam reads to the B. floridae reference genome and we refer these CNEs as cross-species reads mapping-based CNEs (CSRM-based CNEs). The intersection and union of WGA-based CNEs and CSRM-based CNEs sets were also extracted.
