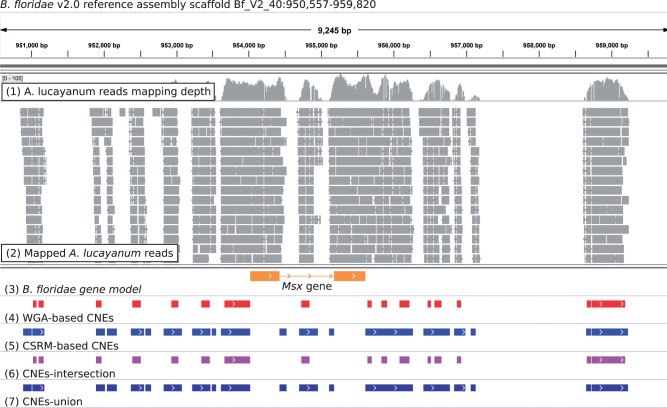
Fig. 2.—
An example of the cephalochordate CNEs identified in this study. The B. floridae Msx gene (B. floridae gene model ID = 278777). The genomic coordinates on the top of the figure show the location (Bf_V2_40:950,557-959,820) of this region according to the B. floridae reference assembly. Seven tracks are shown underneath: 1) A. lucayanum reads mapping depth, 2) mapped A. lucayanum reads, 3) B. floridae gene model, 4) WGA-based CNEs, 5) CSRM-based CNEs, 6) the intersection of WGA-based and CSRM-based CNEs, and 7) the union of WGA-based and CSRM-based CNEs. On the mapped reads track, each gray block represents a mapped A. lucayanum read with mapping quality ≥20. On the B. floridae gene model track, the orange blocks represent the CDS region and the orange line represents the intronic region, whereas the arrows represent the transcription direction of the corresponding gene. On the cephalochordate CNE tracks, each block represents an individual CNE that we identified in this study.