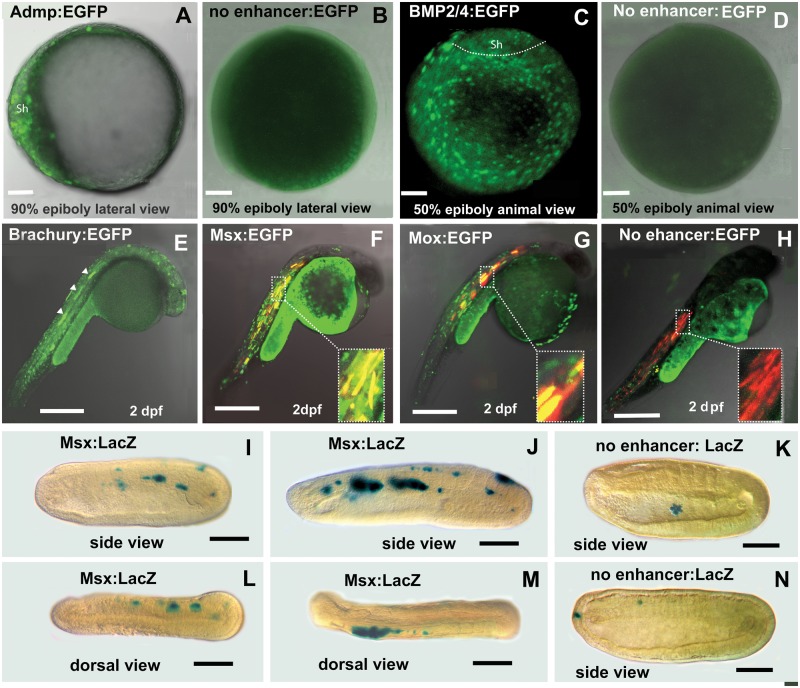
Fig. 4.—
Amphioxus CNEs identified in silico by comparing cephalochordate genomes are functional enhancers in zebrafish. (A) ADMP CNE drives the expression of enhanced green fluorescent protein (EGFP) into dorsal shield of zebrafish late gastrula. (B) Negative control for A shows no expression. (C) EGFP expression driven by BMP2/4 CNE is present throughout the blastoderm with lower intensity in the shield region at gastrula stage. (D) Negative control for C shows no expression. (E) The activity of the Brachyury CNE at late segmentation period. White arrows indicate expression in the notochord. (F) The Msx CNE directs expression to the muscles. Red marks expression of DSRED directed by a muscle-specific zebrafish enhancer. Yellow shows co-expression directed by both enhancers in the muscles. (G) The Mox CNE from amphioxus directs expression to muscle in the zebrafish. Red marks expression directed by a muscle-specific zebrafish enhancer. Yellow shows co-expression directed by both enhancers in the muscles. (H) Negative control. EGFP expression with no enhancer is nonspecific. Red indicates expression of a co-injected muscle-specific enhancer. (I–N). Amphioxus embryos. Anterior to the left. (I) Mid-neurula. Expression driven by the Msx-CNE is mosaic in muscle. (J) Late-neurula. Expression driven by Msx-CNE is expressed strongly in muscle. (K) Negative control. Early neurula. Expression limited to a single sick cell in the gut lumen. (L) Dorsal view. Expression of the Msx-CNE in muscles on the right side. (M) Dorsal view. Expression driven by the Msx-CNE is in muscles on the left side. (N) Negative control. Expression of the empty vector in a single anterior ectodermal cell and in a single cell in the vicinity of the muscles.