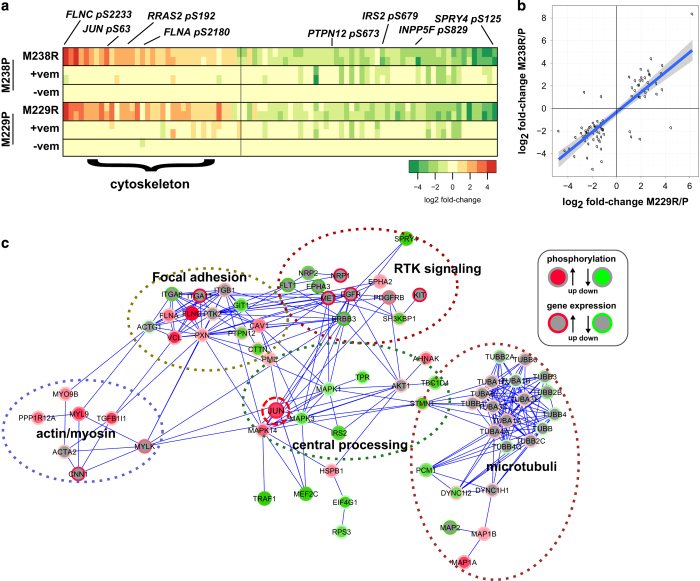
Figure 1.
Proteomic analysis of the global signaling state of melanoma cells with RTK-resistance mechanism reveals extensive alterations in cell attachment and cytoskeleton-related signaling processes. (a) Global phospho-profiling of two parental (P)/resistant (R) cell line pairs of the RTK-resistance mechanism (M229P/R and M238P/R; vem=6 h treatment with 1 μm vemurafenib/PLX4032 (vem)). Each column of the heatmap represents a phospho-peptide. The fold change of phosphorylation compared with the respective parental cell line is color coded in shades of red (upregulated in resistant cells) or green (downregulated in resistant cells). The phospho-profile was generated with a quantitative, label-free mass-spectrometry technique established in our laboratory [38]. (b) Significantly altered phosphorylation sites show same trend in both cell line pairs. A highly significant association between the direction of change for the M229 and M238 pair is observed (Fisher’s exact test; P-value <2.2×10−16). See Supplementary Figure S3 for more details. (c) Illustrative functional protein network of differentially regulated phospho-proteins highlights the observed alterations in cytoskeleton and cell attachment processes in RTK-resistance mechanism cells. The node core and border color shows phosphorylation and gene expression regulation (see legend; darker shades indicate stronger deregulation; missing values are gray). Gene expression data are from Nazarian et al. ([9]; geometric mean of fold changes for M238R/P and M229R/P). The network was generated with the Reactome FI plugin in Cytoscape [94]. The Reactome FI database combines curated interactions from Reactome and other pathway databases with statistically filtered uncurated pairwise relationships from various sources including protein interaction and gene co-expression data sets. Phospho-proteins significantly altered in RTK-resistance mechanism supplemented with differentially regulated genes were linked by functional interactions from the Reactome FI database (network edges). The clusters were identified manually, guided by computational analysis [95].