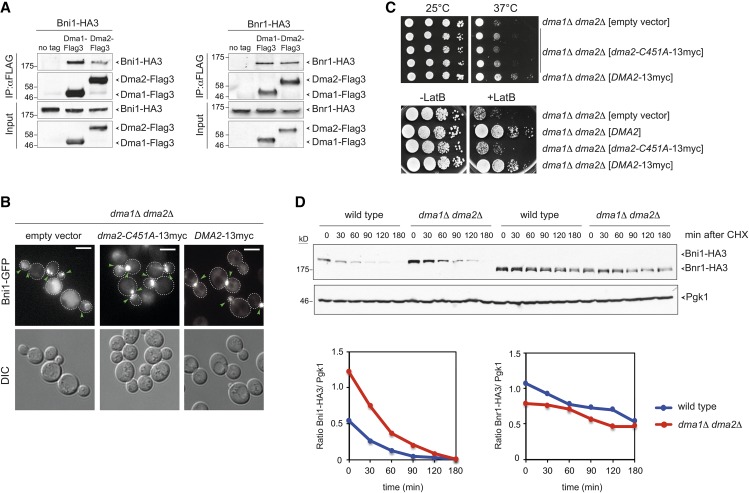
Figure 8.
Dma proteins interact physically with formins and their E3 ligase activity is required for proper Bni1 distribution. (A) Total lysates from logarithmically growing cells expressing at the same time Dma1-Flag3 or Dma2-Flag3 and Bni1-HA3 or Bnr1-HA3 were subjected to immunoprecipitation with anti-Flag antibody followed by Western blot analysis with anti-HA and anti-Flag antibodies. Inputs represent 1/50th of the lysate used for each pull-down. (B) dma1Δ dma2Δ cells expressing Bni1-GFP and carrying a centromeric plasmid to express the catalytically inactive myc-tagged Dma2-C451A (dma2-C451A-13myc) or wild-type Dma2 (DMA2-13myc) were imaged at 25°. Representative images from max-projected Z-stacks (11 planes at 0.3-μm spacing) are shown. Arrowheads indicate different localizations of Bni1. Bar, 5 μm. (C, upper panels) Serial dilutions of cells with the indicated genotypes were spotted on selective (SD −His) plates and incubated at the indicated temperatures. (Lower panels) Serial dilutions of cells with the indicated genotypes were spotted on YEPD either lacking or containing 5 μM LatB and incubated at 25° for 2 days. (D) Wild-type and dma1Δ dma2Δ cells expressing Bni1-HA3 or Bnr1-HA3 were treated with cycloheximide (CHX) at 25° (time 0) to monitor formin degradation by Western blot with anti-HA antibodies. Pgk1 was used as loading control. Ratios between Bni1-HA3 or Bnr1-HA3 levels and the Pgk1 over time were calculated with ImageJ.