Abstract
Glioma constitutes one of the most common groups of brain tumors, and its prognosis is influenced by different genetic and epigenetic modulations. In this study, we demonstrated low or no expression of hedgehog interacting protein (HHIP) in most of the cell lines and primary glioma tumor samples. We further proceeded to promoter methylation study of this gene in the same cell lines and primary tumor samples and found 87 % (7/8) HHIP methylation in glioblastoma cell lines and 75 % (33/44) in primary tumor samples. These methylation pattern correlates with low or unexpressed HHIP in both cell lines and primary tumor samples. Our results suggest the possibility of epigenetic regulation of this gene in glioma, similarly to medulloblastoma, gastric, hepatic, and pancreatic cancers. Also, HHIP might be a diagnostic or prognostic marker in glioma and help to the detection of these tumors in early stages of disease.
Keywords: Human hedgehog interacting protein (HHIP), Glioma, Glioblastoma promoter methylation, Melting curve analysis-Meth (MCA-Meth), GLI1
Introduction
Glioma is the most common worldwide reported brain tumor. In spite of intensive research on tumor biology, the application of these researchers’ yields to improve the survival of glioma patients is still a challenge for oncologists. The major influencing forces for this unsuccessful cure of glioma are due to the involvement of both known and unknown genes in the genesis of glioma. Even if we could understand some of the molecular mechanisms of gene regulation in glioma development, still, the influence of the environment on these tumors would remain a milestone for cancer biologists. Therefore, it would be better to apply holistic approaches of biomedical research which could cover all possible factors influencing this kind of malignant transformation.
It has been reported that many signaling pathways are involved in brain development and among these pathways, sonic hedgehog has a significant contribution for neurogenesis [1]. Sonic hedgehog signaling has both mitogenic and morphogenic characters [2, 3]. This signaling pathway starts from the binding of the N-terminal cholesterol modified Shh protein to the twelve transmembrane PTCH1 receptor. Before the binding of Shh ligand to PTCH1, this PTCH1 inhibits another seven transmembrane receptor, Smoothened (SMO), of this pathway. After binding of Shh ligand to PTCH1, SMO is relieved, which further enters in the cytoplasm and activate a main transcriptional factor zinc finger protein, GLI1, with the help of serine/threonine proteins [4, 5]. The mechanism of PTCH1 and SMO interaction is not too clear; however, there is speculation that some conformational changes occur to relieve SMO from PTCH1 inhibition. This activates GLI1, which now moves from the cytoplasm to the nucleus for the regulation of Shh signaling target genes including PTCH1, cyclin D, plakoglobin, and many more [6].
Importantly, PTCH1 is one of the downstream target genes of Shh signaling but also acts as a negative regulator of this signaling pathway activation. Similarly, another receptor named hedgehog interacting protein (HHIP) binds to all three hedgehog proteins namely SHH, IHH, DHH and inhibits their downstream signaling activation. The binding affinity of HHIP to Shh is not less than to PTCH1; therefore, both HHIP and PTCH1 act as competitors for binding to Shh ligands [7]. HHIP is also one of the downstream target genes of Shh signaling and acts as a tumor suppressor gene [8]. It is well known that Shh signaling participates in early development, but Shh is also involved in cancer initiation and/or progression. Since HHIP is a negative regulator of Shh signaling activation, the possibility of genetic and epigenetic alteration of this gene in malignant transformation has been increased. Indeed, the absence or low expression of HHIP in Shh activated cell lines and samples of pancreatic [9], hepatic [10] and gastric cancer [11], and medulloblastoma [12], has been documented. Low expression of HHIP further restored in pancreatic, gastrointestinal, and hepatic cancer cell lines after treatments with the demethylating agent 5-aza-2′-deoxycytidine. These results confirm the promoter methylation of HHIP in these cancers. Nevertheless, other reports showed histone deacetylation and chromatin remodeling contributing to silencing of HHIP expression in gastrointestinal and pancreatic cancer [11, 13]. These studies indicate the epigenetic regulation of HHIP in various cancers.
In this study, we have checked the messenger RNA (mRNA) expression of HHIP gene in 8 glioma cell lines and 27 primary tumors samples. Our transcript expression results further led us to analyze the promoter of HHIP gene in all 8 cell lines and 44 primary tumor samples. However, our promoter methylation analysis pattern was different from conventional methylation-specific PCR (MSP). For promoter methylation analysis, we applied melting curve analysis-Meth (MCA-Meth) assay of one pair of primers. This technique gave us not only accurate percentage of methylation in the promoter region but also a very easy to check assay for methylation in large cohorts of patients in a short time.
Materials and methods
Cell lines
We used 8 glioma cell lines, namely U87MG, A172, LN405, SW1783, T98G, SW1088, CCF-STTG-1, and Gos-3. Cell lines U87MG, A172, and T98G were purchased from the European Collection of Cell Cultures (Salisbury, Wiltshire, UK). Cell lines SK-PN-DW, CCF-STTG1, SW1088, and SW1783 were purchased from the American Type Culture Collection (Manassas, VA, USA). Cell lines LN405 and Gos-3 were obtained from the Deutsche Sammlung Von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Cell lines were cultured in RPMI L-Glutamax medium (GIBCO-BRL, Gaithersburg, MD, USA), supplemented with 10 % fetal bovine serum (FBS), 10 % non-essential amino acids (NEAs), 1 % penicillin, and 0.1 % amphotericin B. Cell lines were maintained in a 37 °C incubator with the supply of 5 % CO2. Sub-culturing of cells was performed after 80 % confluence with the help of trypsin/EDTA 1X.
Primary tumor samples
We used 27 glioma primary tumor samples for expression analysis and 44 samples for promoter methylation study. All samples were collected from the Hospital of Navarra, Pamplona, Spain. The use of these samples was approved by the Ethical Committee of the University of Navarra, under protocols 38/2002 and 83/2011.
qRT-PCR
RNA was extracted from 8 glioma cell lines by using the Quick Prep Total RNA extraction Kit (Amersham Bioscience, UK). We purchased normal brain tissue RNA (Stratagene, Cedar Creek, TX) for normal control expression. A total of 1 μg of RNA was converted into complementary DNA (cDNA) by Superscript II Rnase H Reverse Transcriptase kit (Invitrogen, Life Technologies, Carlsbad, CA). These converted cDNAs were taken for expression study of HHIP and GLI1 [14] with the help of a Bio-Rad iQ 5 qRT-PCR machine. All samples were run in triplicate, and their expression results were normalized with housekeeping gene GAPDH. The primer sequences for HHIP gene were forward 5′ATGGTGGGTT GTGCTTTCC3′, reverse 5′AGTTGTGTTTGTGCTTTCTG CT3′, and for GLI1 gene, as in a previous study [14]. The reaction conditions for qRT-PCR were denaturation at 95 °C for 5 min, then 35 cycles of denaturation (95 °C for 1 min), annealing (59.5 °C for 40 s), and extension (72 °C for 50 s); and final extension at 72 °C for 10 min; then further melting curve analysis at 72 °C for 1 min, and finally 95 °C for 10 min.
Promoter analysis of HHIP
We selected HHIP promoter sequences from a previous published report [15]. We further explored the possible CpG islands in this 1501-bp promoter with the help of Methprimer [16]. We identified two putative CpGs rich regions of 373 and 224 bp; they were considered first and second CpG islands, respectively [12]. The evaluation of the promoter’s CpG islands was based on standard criteria, island size>200 bp, GC%>50, Obs/Exp>0.6 [12].
DNA extraction and bisulphite modification
DNA was extracted from the 8 cell lines and the 44 primary tumor samples by the Wizard Genomic DNA Purification Kit (Promega, Madrid, Spain), according to manufacturer’s protocol. We used a total of 1-μg DNA for bisulphite modification with CpGenome™ DNA Modification Kit (Chemicon International, Darmstadt, Germany). Normal blood DNA and in vitro methylated DNA-Genome Universal Methylated DNA (Chemicon International, Darmstadt, Germany) were considered as negative and positive controls of promoter methylation analysis, respectively.
Methylation assay (MCA-Meth)
Melting curve analysis-Meth (MCA-Meth) was set up in our laboratory [17]. In MCA-Meth method, designed primers are free from CpG sequences and amplify the target region unbiased. This technique is suitable and efficient for the high-throughput analysis of heterogeneous pools of methylation in cell lines and primary tumor samples. The method differentiates methylated and unmethylated DNA at the gene promoter, on the basis of melting curve and melting temperature. For MCA-Meth, 5 ng of bisulphite DNA is mixed with 2.5 pmol of forward and reverse primers in 2× IQ SYBR Green supermix (Bio-Rad, Hercules, CA, USA) up to 25 μl of total reaction volume. The PCR reaction conditions were 94 °C for 10 min, then 30 cycles of denaturation (94 °C for 30 s), annealing (64 °C for 40 s), and extension (72 °C for 30 s); and final extension at 72 °C for 10 min. Melting curve analysis was performed between 70 to 90 °C, and temperature was increased by 0.5 °C every 30 s. DNA amplification and melting curve analysis were done in an iQ5 Multicolor Real Time PCR Detection System (Bio-Rad, Hercules, CA, USA).
Statistical analysis
All the expression study was performed in triplicate, including SD calculations.
Results
Expression of HHIP in glioma cell lines and tumor samples
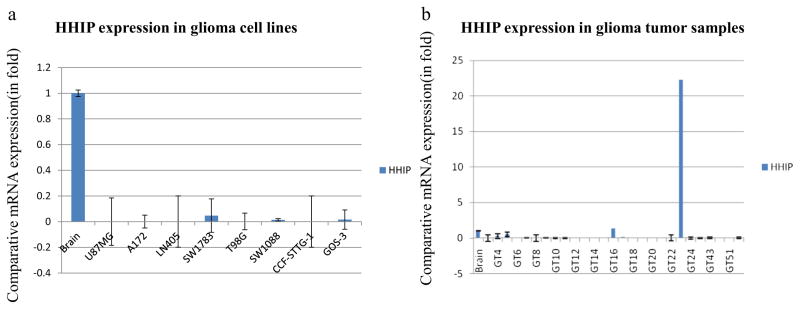
We checked the mRNA expression of the HHIP gene in 8 glioma cell lines. Interestingly, most of the cell lines either showed low expression (SW1783, SW1088, and Gos-3) or no expression (U87MG, A172, LN405, T98G, and CCF-STTG-1) in comparison to normal brain tissue (Fig. 1a). Similarly, we checked the expression of HHIP in 27 primary tumor samples and found that most samples showed either low or no expression compared to normal brain tissue (Fig. 1b) (Tables 1 and 2).
Fig. 1.
Comparative HHIP mRNA expression in glioma cell lines and tumor samples. a HHIP expression in glioma cell lines. b HHIP expression in primary tumor samples
Table 1.
HHIP expression and promoter methylation pattern in glioma cell lines
| Cell lines | HHIP expression | HHIP promoter methylation | % of promoter methylation |
|---|---|---|---|
| U87MG | ± | U+M | 60–65 |
| A172 | ± | M | 90–95 |
| LN405 | ± | U+M | 60–65 |
| SW1783 | −/+ | U | 0 |
| T98G | ± | U+M | 60–65 |
| SW1088 | ± | U+M | 50–55 |
| CCF-STTG | ± | M | 90–95 |
| GOS-3 | −/+ | M | 90–95 |
“±” no detectable expression, “−/+” low expression
U unmethylation, M methylation, U+M hemi/partial methylation
Table 2.
HHIP expression and promoter methylation pattern in glioma samples
| Sample No. | Samples ID | Expression | Methylation by melting curve | % of methylation |
|---|---|---|---|---|
| 1 | GT-3(GBM) | ± | M | 85–90 |
| 2 | GT-4(AIII) | + | U+M | 60–65 |
| 3 | GT-5(GBM) | + | U | 0 |
| 4 | GT-6(GBM) | – | M | 85–90 |
| 5 | GT-7(AIII) | – | M | 85–90 |
| 6 | GT-8(GBM) | – | U+M | 55–60 |
| 7 | GT-9(GBM) | ± | U+M | 55–60 |
| 8 | GT-10(AIII) | ± | U+M | 55–60 |
| 9 | GT-11(GBM) | – | nd | nd |
| 10 | GT-12(GBM) | – | nd | nd |
| 11 | GT-13(GBM) | ± | U+M | 60–65 |
| 12 | GT-14(AIII) | −/+ | M | 85–90 |
| 13 | GT-15(GBM) | −/+ | U+M | 55–60 |
| 14 | GT-16(AIII) | + | M | 80–90 |
| 15 | GT-17(GBM) | + | M | 80–90 |
| 16 | GT-18(GBM) | −/+ | nd | nd |
| 17 | GT-19(AIII) | – | U+M | 55–60 |
| 18 | GT-20(AII) | – | U+M | 55–60 |
| 19 | GT-21(AIII) | – | U+M | 60–65 |
| 20 | GT-22(GBM) | ± | M | 85–90 |
| 21 | GT-23(GBM) | – | M | 85–90 |
| 22 | GT-24(GBM) | ± | U+M | 55–60 |
| 23 | GT-25(AI) | nd | U | 0 |
| 24 | GT-26(GBM) | nd | M | 85–90 |
| 25 | GT-27(GBM) | nd | M | 85–90 |
| 26 | GT-28(GBM) | nd | nd | nd |
| 27 | GT-29(GBM) | nd | M | 85–90 |
| 28 | GT-30(GBM) | −/+ | M | 85–90 |
| 29 | GT-31(GBM) | nd | M | 90–95 |
| 30 | GT-32(GBM) | nd | U+M | 55–60 |
| 31 | GT-34(AI) | nd | U+M | 55–60 |
| 32 | GT-35(AIII) | nd | U+M | 60–65 |
| 33 | GT-36(GBM) | nd | U+M | 60–65 |
| 34 | GT-41(GBM) | nd | U | 0 |
| 35 | GT-43(GBM) | −/+ | U+M | 60–65 |
| 36 | GT-44(GBM) | nd | U+M | 60–65 |
| 37 | GT-45(GBM) | nd | U+M | 40–45 |
| 38 | GT-46(GBM) | nd | U+M | 60–65 |
| 39 | GT-47(GBM) | nd | U | 0 |
| 40 | GT-48(GBM) | nd | nd | nd |
| 41 | GT-49(GBM) | nd | U+M | 50–55 |
| 42 | GT-50(AII) | – | U+M | 50–55 |
| 43 | GT-51(GBM) | ± | nd | nd |
| 44 | GT-52(GBM) | ± | nd | nd |
“–” no expression, “±” no detectable expression, “−/+” low expression, “+”expression, “nd” not determined
U unmethylation, M methylation, U + M hemi/partial methylation
Promoter methylation analysis of HHIP in glioma cell lines
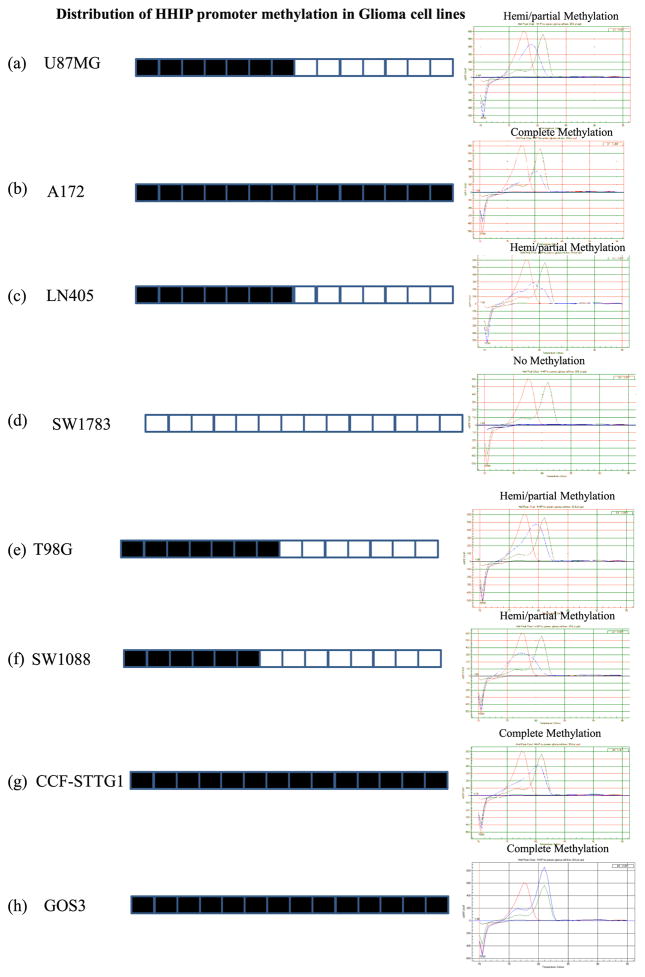
Low expression of HHIP mRNA among cell lines and tumor samples led us further into exploring the possibility of promoter methylation in these cell lines and tumor samples. We performed promoter methylation analysis with MCA-Meth assay. In this method, we successfully analyzed the methylation pattern of HHIP promoter in 8 glioma cell lines and 44 primary tumor samples. Three glioma cell lines (A172, CCF-STTG-1, and Gos-3) showed complete methylation (90–95 % methylation) (Fig. 2b, g, and h), while four cell lines (U87MG, LN405, T98G, and SW1088) showed partial/hemi methylation (50–65 %) (Fig. 2a, c, e, and f) (Table 1). However, we were unable to determine the methylation status of one cell line SW1783 (Fig. 2d). Interestingly, all of these glioma cell lines showed low fold expression of HHIP transcript compared to normal brain tissue. Our amplified product of this methylation study contains a total of 14 CpGs, and on the basis of number of CpGs methylated in the cell lines or tumor samples, we consider them either complete methylation or hemi/partial methylation (Fig. 2).
Fig. 2.
HHIP promoter with 14 CpGs and methylation pattern distribution by MCA-Meth in glioma cell lines. a U87MG shows hemi/partial methylation (60–65 %). b A172 shows complete methylation (90–95 %). c LN405 shows hemi/partial methylation (60–65 %). d SW1783 shows no methylation. e T98G shows hemi/partial methylation (60–65 %). f SW1088 shows hemi/partial methylation (50–55 %). g CCF-STTG1 shows complete methylation (90–95 %). h GOS3 shows complete methylation (90–95 %)
Promoter methylation analysis of HHIP in glioma primary tumor samples
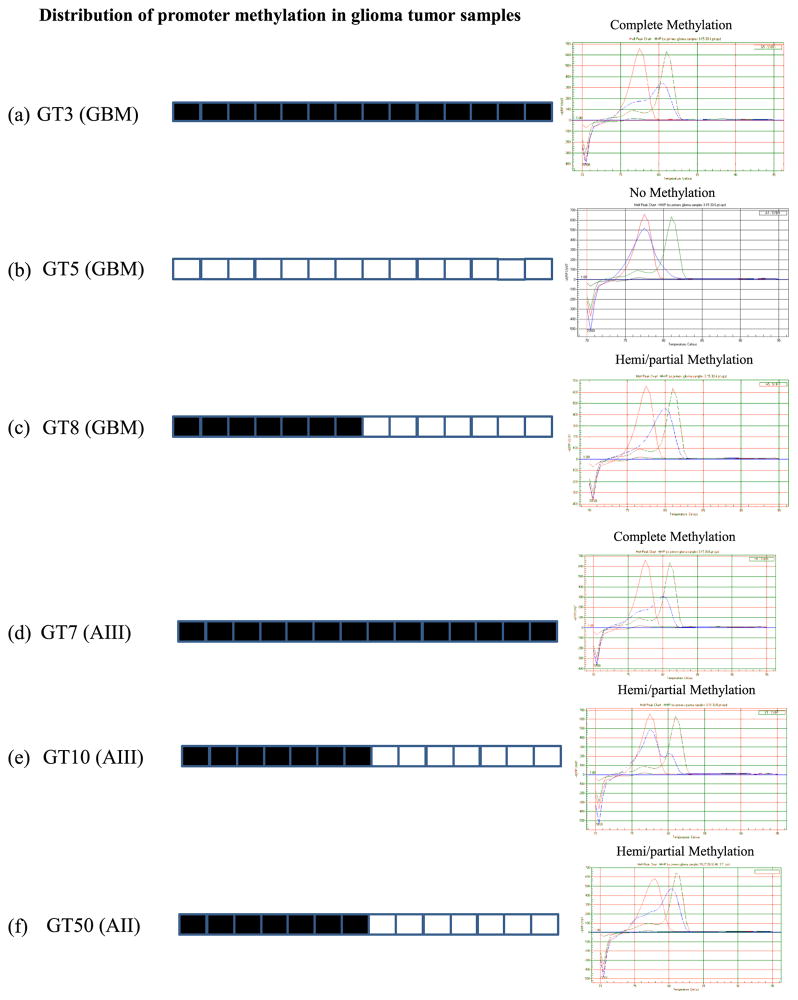
Similarly, we applied the MCA-Meth method for HHIP promoter methylation analysis in 44 primary tumor samples. Among 44 primary tumor samples, 75 % (33/44) samples showed methylation, including complete methylation (14/ 44) and hemi/partial methylation (19/44), and 14 % (6/44) samples showed no methylation in the HHIP promoter (Fig. 3, Table 2). We were unable to determine methylation in the remaining 11 % (5/44) tumor samples (Fig. 3, Table 2).
Fig. 3.
HHIP promoter with 14 CpGs and methylation pattern distribution by MCA-Meth in glioma tumor samples. Percentage of methylation varies from no methylation (0 %) to complete methylation (90–95 %). a GT3 (GBM) shows complete methylation (90–95 %). b GT5 (GBM) shows no methylation. c GT8 (GBM) shows hemi/partial methylation (55–60 %). d GT7 (AIII) shows no methylation. e GT10 (AIII) shows hemi/partial methylation (55–60 %). f GT50 (AII) shows hemi/partial methylation (50–55 %). AII low-grade (fibrillary) astrocytoma (grade II), AIII anaplastic astrocytoma (grade III), GBM glioblastoma multiforme (grade IV)
Discussion
Glioma is a major tumor of the brain, and in general, prognosis is bad because there is not any defined identity of prominent and specific biomarkers of this tumor in its early stage of development. Later therapies remain ineffective due to the multiple causative factors that are involved in the progression of this malignant tumor. Neurooncologists are trying to minimize the side effects of glioma therapies and to enhance survival of cancer patients.
Shh is a major signaling pathway that shows both epigenetic and genetic alteration lading to malignant transformation of various normal tissues into different types of cancers [18–23]. Mutation of the SMO and PTCH1 receptors promote basal cell carcinoma and medulloblastoma formation, while other studies also suggest that the activation of this pathway has been defined by the GLI1 marker, being its high expression and indication of the possibility of Shh activation [14, 24–27].
The role of HHIP has been defined as antagonist of Shh signaling and shows similar affinity as PTCH1 for binding to three homologous of hedgehog proteins SHH, IHH, and DHH. Low expression of HHIP has been reported in cancers like pancreatic [9], gastrointestinal [11], hepatic [10, 28], or medulloblastoma [12]. Epigenetic modification of tumor suppressor genes in cancer progression is well known. HHIP is considered to be a tumor suppressor gene. To our knowledge, there is not any report so far on HHIP expression and promoter methylation in glioma. We attempted to explore the expression of this gene and its possible promoter hyper-methylation in glioma cell lines and primary tumor samples. Our results support the fact that low or no expression of HHIP associates to promoter methylation in glioma cell lines. So far, we found two CpGs rich regions in the span of 1500 bases of promoter after analysis of HHIP promoter regions with Meth primers [16]; the first CpG island is 373 bp in length and the second one extends along 224 bp (CpG islands were selected under standard criteria). Our primers for methylation analysis in the first CpG island produced an amplified region of 227 bp with 14 CpGs. Maybe some other factors might influence the expression of the second CpG island. This promoter is known to have 11 consensus sequences for the binding of bHLA transcription factor, which is downregulated by Shh signaling in vascular remodeling [15].
As far as we know from the literature, not only promoter methylation leads to silencing of HHIP gene expression in pancreatic cancer. On the contrary, several other epigenetic factors need to be explored [9]. In another study, it has been reported that not only epigenetic alterations causes silencing of HHIP gene but also many other factors like loss of heterozygosity, somatic cell mutation, and also regulation of microRNA in human hepatocellular carcinoma [10]. Interestingly, besides HHIP promoter, H3-K4 and H3-K9 methylation also contribute to silencing of HHIP in gastrointestinal cancer [11].
Among tumor samples, only 33/44 (75 %) samples showed complete or hemi/partial methylation and 6/44 (14 %) samples did not show methylation based on their melting curve analysis. But most tumor samples did not show expression of HHIP. This discrepancy suggests two possibilities: either there is methylation in the second promoter region, or other factors may be involved in HHIP expression including signaling pathways like sonic hedgehog, notch or both, suppressing the expression of this gene [8, 15].
Taken together, our results of HHIP expression and methylation analysis in glioma cell lines and primary tumors indicate the possibility of HHIP epigenetics, but also an activated Shh signaling pathway that downregulates HHIP in glioma. This possibility is also supported by previous studies which show the contribution of Shh signaling in glioma and the development of other tumors [24–26, 29].
Acknowledgments
M.H. Shahi was a fellow of AECI (Agencia Española de Cooperación Internacional), Madrid, Spain. This research was supported in part by grants from the Departamento de Salud del Gobierno de Navarra, Caja Navarra (project 13912), Fundación Universitaria de Navarra, Pamplona, and Fondo de Investigación Sanitaria (PI-081849, to JSC and PI13/00055, to JAR), Madrid.
Contributor Information
Mehdi H. Shahi, Brain Tumor Biology Unit, University of Navarra School of Sciences, Pamplona, Spain
Idoya Zazpe, Neurosurgery Service, Hospital of Navarra, Pamplona, Spain.
Mohammad Afzal, Department of Zoology, Aligarh Muslim University, Aligarh, India.
Subrata Sinha, National Brain Research Centre, Manesar, Gurgaon, India.
Robert B. Rebhun, Department of Surgical and Radiological Sciences, University of California Davis School of Veterinary Medicine, Davis, CA, USA
Bárbara Meléndez, Molecular Pathology Research Unit, Department of Pathology, Virgen de la Salud Hospital, Toledo, Spain.
Juan A. Rey, IdiPaz Research Unit, La Paz University Hospital, Madrid, Spain
Javier S. Castresana, Email: jscastresana@unav.es, Brain Tumor Biology Unit, University of Navarra School of Sciences, Pamplona, Spain
References
- 1.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 2.Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol. 2004;166:673–84. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–83. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 4.Preat T, Therond P, Lamour-Isnard C, Limbourg-Bouchon B, Tricoire H, Erk I, et al. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature. 1990;347:87–9. doi: 10.1038/347087a0. [DOI] [PubMed] [Google Scholar]
- 5.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–45. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 6.Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA, et al. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277:5548–55. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- 7.Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–7. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–21. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 9.Martin ST, Sato N, Dhara S, Chang R, Hustinx SR, Abe T, et al. Aberrant methylation of the Human Hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer Biol Ther. 2005;4:728–33. doi: 10.4161/cbt.4.7.1802. [DOI] [PubMed] [Google Scholar]
- 10.Tada M, Kanai F, Tanaka Y, Tateishi K, Ohta M, Asaoka Y, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14:3768–76. [Google Scholar]
- 11.Taniguchi H, Yamamoto H, Akutsu N, Nosho K, Adachi Y, Imai K, et al. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. J Pathol. 2007;213:131–9. doi: 10.1002/path.2216. [DOI] [PubMed] [Google Scholar]
- 12.Shahi MH, Afzal M, Sinha S, Eberhart CG, Rey JA, Fan X, et al. Human hedgehog interacting protein expression and promoter methylation in medulloblastoma cell lines and primary tumor samples. J Neurooncol. 2011;103:287–96. doi: 10.1007/s11060-010-0401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun SG, Zhou W, Yee NS. Combined targeting of histone deacetylases and hedgehog signaling enhances cytoxicity in pancreatic cancer. Cancer Biol Ther. 2009;8:1328–39. doi: 10.4161/cbt.8.14.8633. [DOI] [PubMed] [Google Scholar]
- 14.Shahi MH, Lorente A, Castresana JS. Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol Rep. 2008;19:681–8. [PubMed] [Google Scholar]
- 15.Katoh Y, Katoh M. Comparative genomics on HHIP family orthologs. Int J Mol Med. 2006;17:391–5. [PubMed] [Google Scholar]
- 16.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 17.Lorente A, Mueller W, Urdangarin E, Lazcoz P, von Deimling A, Castresana JS. Detection of methylation in promoter sequences by melting curve analysis-based semiquantitative real time PCR. BMC Cancer. 2008;8:61. doi: 10.1186/1471-2407-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Utikal J, Gratchev A, Muller-Molinet I, Oerther S, Kzhyshkowska J, Arens N, et al. The expression of metastasis suppressor MIM/MTSS1 is regulated by DNA methylation. Int J Cancer. 2006;119:2287–93. doi: 10.1002/ijc.22106. [DOI] [PubMed] [Google Scholar]
- 19.Wang LH, Choi YL, Hua XY, Shin YK, Song YJ, Youn SJ, et al. Increased expression of sonic hedgehog and altered methylation of its promoter region in gastric cancer and its related lesions. Mod Pathol. 2006;19:675–83. doi: 10.1038/modpathol.3800573. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Yang X, Li J, Tian X, Cai J, Zhang Y. Opposite expression patterns of Sonic hedgehog and Indian hedgehog are associated with aberrant methylation status of their promoters in colorectal cancers. Pathology. 2010;42:553–9. doi: 10.3109/00313025.2010.508785. [DOI] [PubMed] [Google Scholar]
- 21.Shahi MH, Schiapparelli P, Afzal M, Sinha S, Rey JA, Castresana JS. Expression and epigenetic modulation of sonic hedgehog-GLI1 pathway genes in neuroblastoma cell lines and tumors. Tumour Biol. 2011;32:113–27. doi: 10.1007/s13277-010-0105-x. [DOI] [PubMed] [Google Scholar]
- 22.Shahi MH, Afzal M, Sinha S, Eberhart CG, Rey JA, Fan X, et al. Regulation of sonic hedgehog-GLI1 downstream target genes PTCH1, Cyclin D2, Plakoglobin, PAX6 and NKX2.2 and their epigenetic status in medulloblastoma and astrocytoma. BMC Cancer. 2010;10:614. doi: 10.1186/1471-2407-10-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TP, Hsu SH, Feng HC, Huang RF. Folate deprivation enhances invasiveness of human colon cancer cells mediated by activation of sonic hedgehog signaling through promoter hypomethylation and cross action with transcription nuclear factor-kappa B pathway. Carcinogenesis. 2012;33:1158–68. doi: 10.1093/carcin/bgs138. [DOI] [PubMed] [Google Scholar]
- 24.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 26.Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–2. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 27.Zurawel RH, Allen C, Chiappa S, Cato W, Biegel J, Cogen P, et al. Analysis of PTCH/SMO/SHH pathway genes in medulloblastoma. Genes Chromosomes Cancer. 2000;27:44–51. doi: 10.1002/(sici)1098-2264(200001)27:1<44::aid-gcc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 28.Eichenmuller M, Gruner I, Hagl B, Haberle B, Muller-Hocker J, von Schweinitz D, et al. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49:482–90. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 29.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]