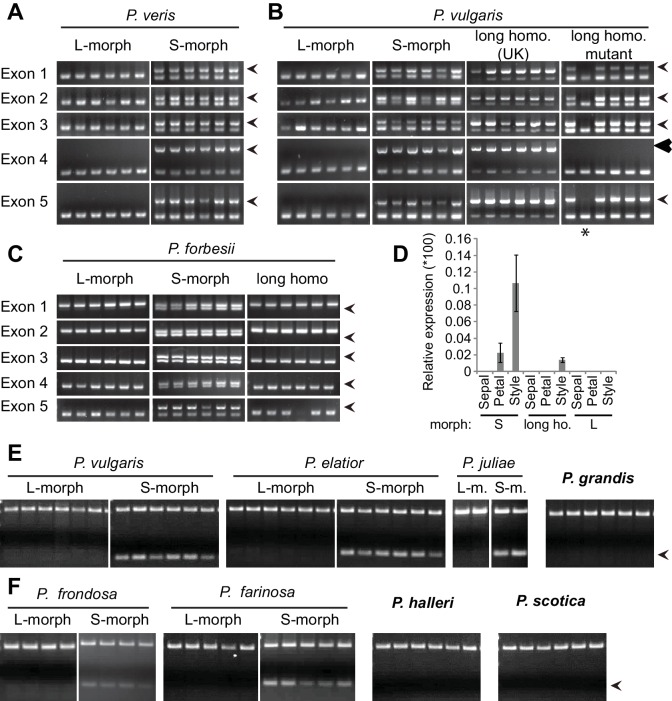
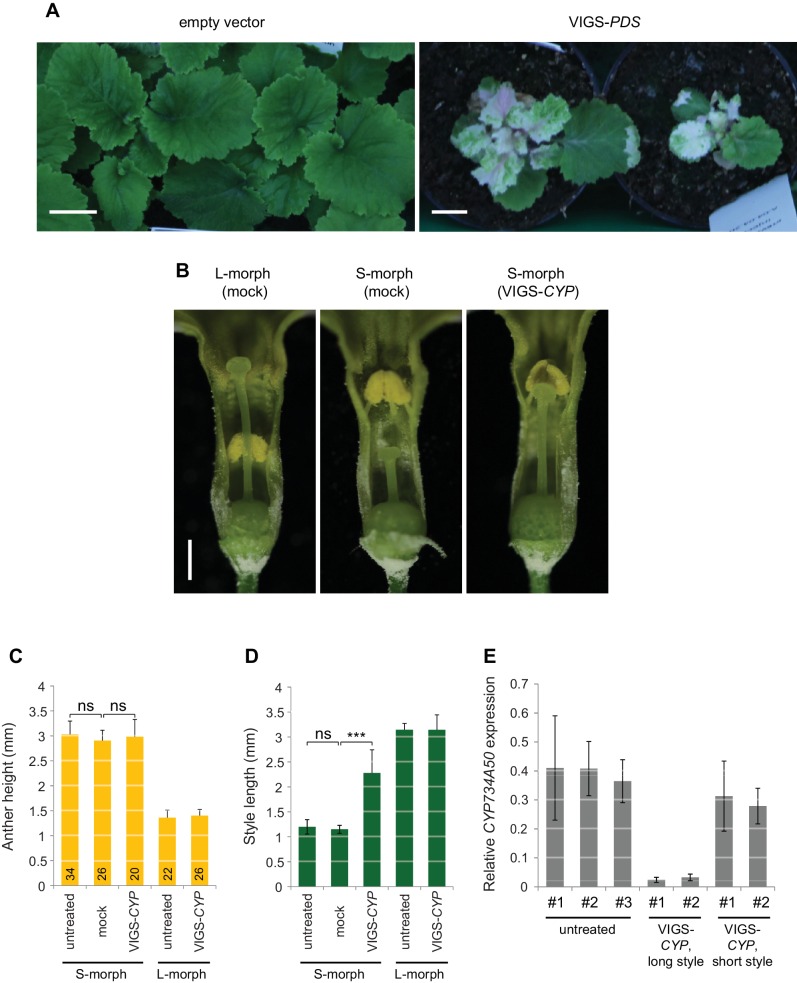
Figure 2. Co-segregation of CYP734A50 with the style phenotype.
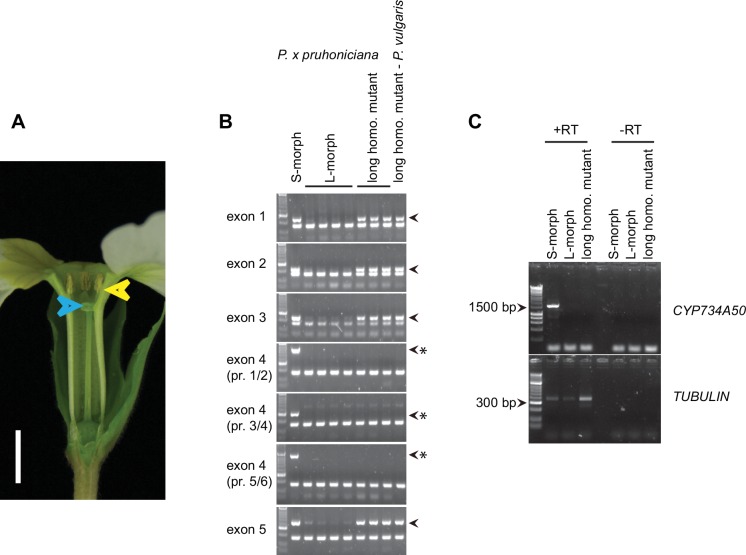
(A–C) PCR-genotyping of individuals with the indicated phenotypes of P. veris (A), P. vulgaris (B) and P. forbesii (C). ‘Long homo (UK)’ are naturally occurring long homostyles from England; ‘long homo mutant’ are long homostyles from the commercially grown population. ‘Long homo’ P. forbesii are long-homostylous putative recombinants found in our experimental population. Multiplex PCR amplified CYP734A50 exons (arrowheads) and ITS as an internal control. Enlarged arrowhead marks absence of exon 4 in the mutant. Asterisk in (B) marks a segregating L-morph individual. (D) Expression of CYP734A50 in dissected floral organs of naturally occurring long homostyles, S- and L-morphs of P. vulgaris, normalized to α-TUBULIN. Values are mean ± SD from three biological replicates. (E,F) PCR-genotyping of individuals with the indicated phenotypes from species in Primula sect. Primula (E) and Primula sect. Aleuritia (F). Long homostylous species are indicated in bold. Multiplex PCR was performed using primers to amplify exon 3 of CYP734A50 (arrowhead) and ITS as an internal control. Full gel images are shown in Figure 2—figure supplement 2. Specificity of PCR amplification for CYP734A50 is shown in Figure 2—figure supplement 3. Each set of plants was genotyped at least twice independently.
DOI: http://dx.doi.org/10.7554/eLife.17956.008
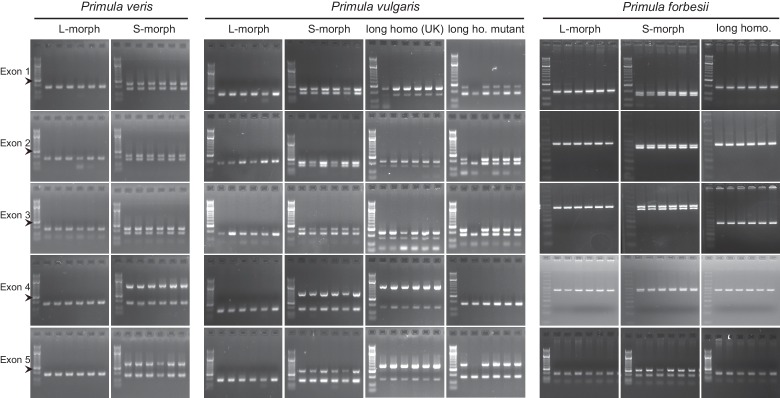
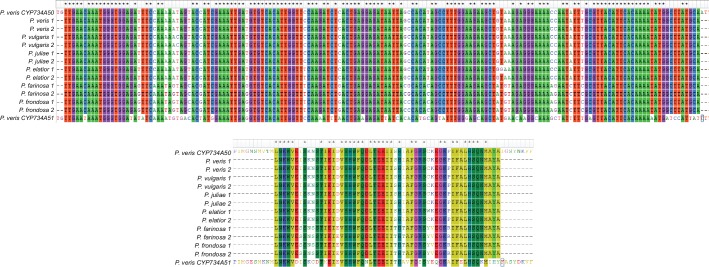
Figure 2—figure supplement 1. Co-segregation of CYP734A50 with the style phenotype.
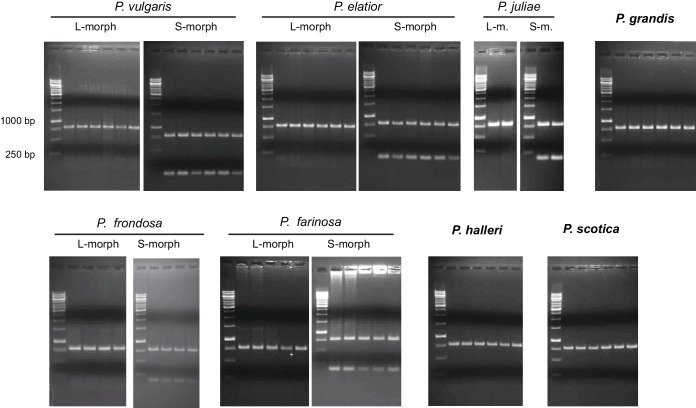
Figure 2—figure supplement 2. Loss of CYP734A50 from natural long homostylous species.
Figure 2—figure supplement 3. Specificity of PCR-genotyping in different Primula species.
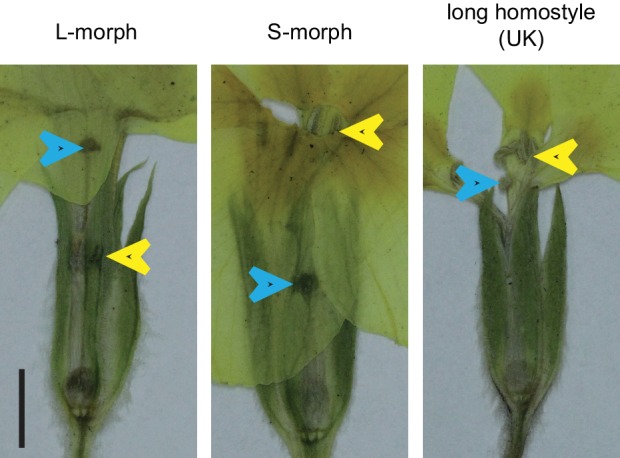
Figure 2—figure supplement 4. Naturally occurring long homostyles of P. vulgaris.