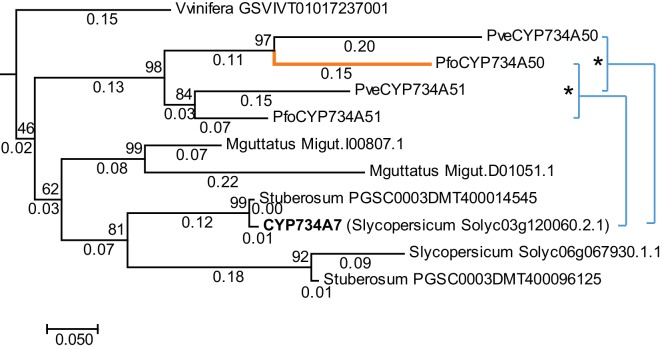
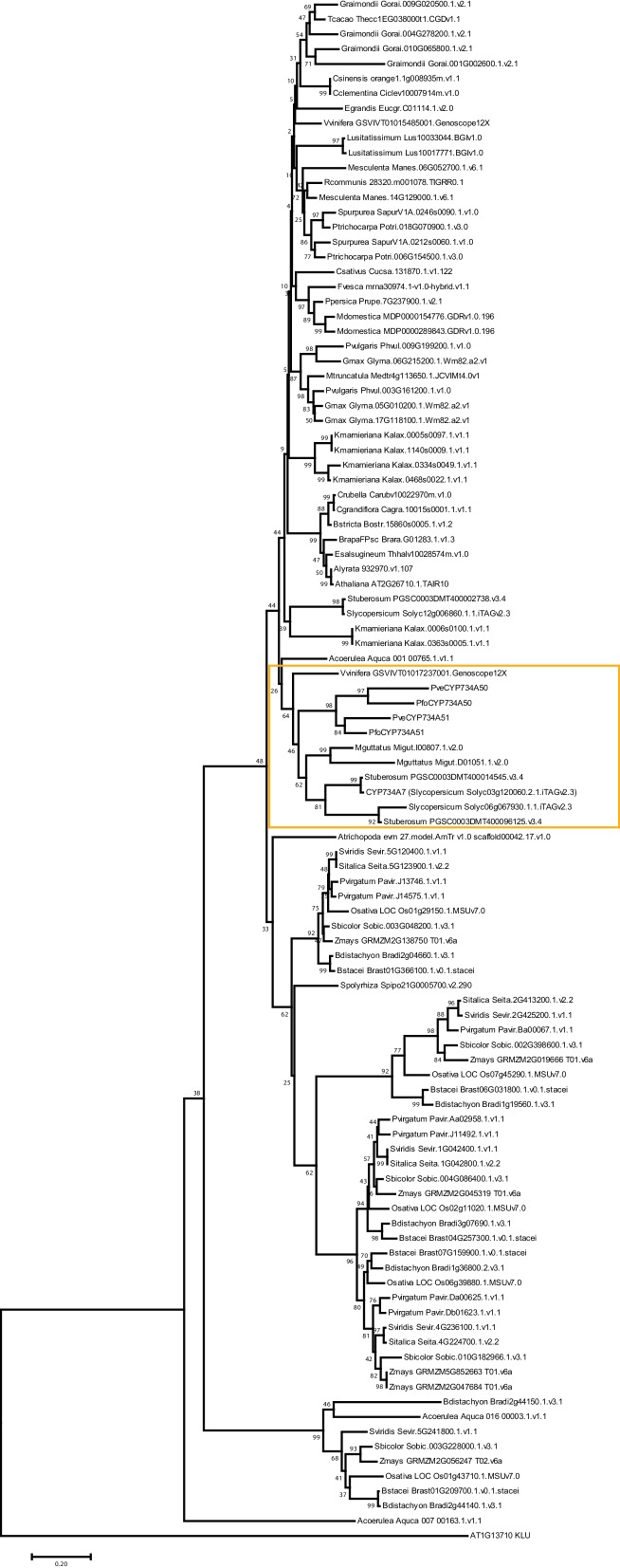
Figure 4. Phylogenetic analysis of CYP734A-like proteins.
Molecular phylogeny of CYP734A50-related proteins based on Neighbour Joining. The tree is drawn to scale, with branch lengths (below branches) measured as number of substitutions per site. Numbers next to nodes indicate percent support from bootstrap analysis (1000 replicates). Cyan brackets with asterisks show significant differences in evolutionary rates compared to CYP734A7 (bold) as outgroup as assessed by Tajima’s relative rate test (p<0.01). CYP734A7 causes BR-deficient phenotypes when overexpressed (Ohnishi et al., 2006). The orange branch shows evidence for relaxed constraint, but not for positive selection based on branch-site tests 1 and 2. The full tree is shown in Figure 4—figure supplement 1. Full results of branch-site tests 1 and 2 are shown in Figure 4—source data 2.
DOI: http://dx.doi.org/10.7554/eLife.17956.018