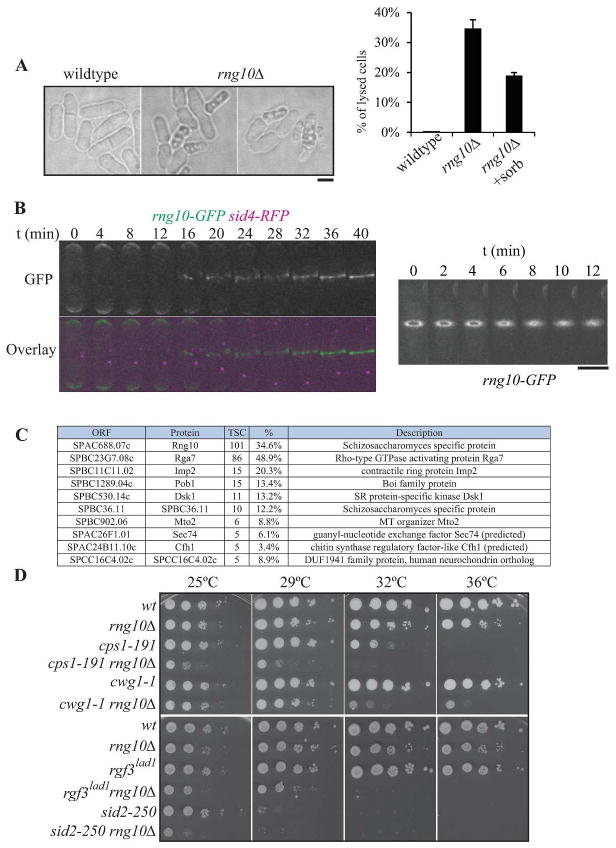
Figure 1. Characterization of SPAC688.07cΔ (rng10Δ).
A) Differential interference contrast (DIC) images of wildtype and rng10Δ cells, illustrating the cell lysis phenotype. Quantitation of cell lysis is included to the right (n=100 for each data point, the average from three replicates and standard deviation are plotted). Scale bar, 5 μm. B) Montages of live cell images of rng10-GFP sid4-RFP cells. Left panel emphasizes the arrival time of Rng10 to the division site; right panel shows Rng10 localization during contractile ring constriction. Scale bar for both right and left panels, 5 μm. C) List of proteins that localize to the cell division site identified by LC-MS of a Rng10-GFP GFP-trap experiment. TSC = total spectral counts. D) Negative genetic interactions of rng10Δ with mutations in glucan synthase genes (cps1/bgs1 and bgs4/cwg1) and septation regulatory genes (rgf3 and sid2). 10-fold dilutions of the indicated strains were spotted on YE plates and incubated for 2–3 days at the indicated temperatures.