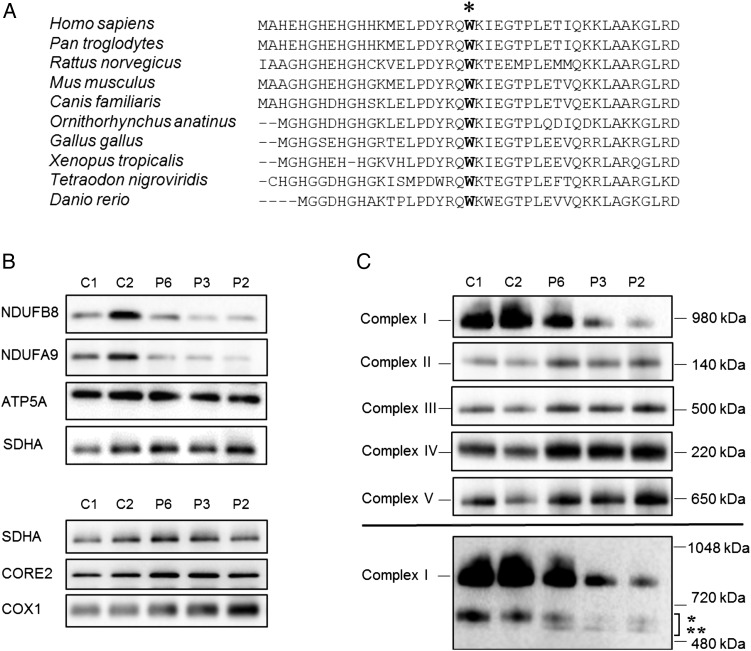
Figure 2.
Analysis of OXPHOS complex assembly and protein expression levels (A) Clustal Omega sequence alignment shows the evolutionary conservation of the p.Trp22 residue (marked with asterix), based on the human sequence (amino acids 1–43). (B) Immunoblot analysis of steady state levels of OXPHOS subunits in mitochondrial lysates isolated from control (C1, C2) and patient skeletal muscle samples (P6, P3, P2). OXPHOS subunit-specific antibodies against the indicated proteins showed a marked decrease in Complex I subunits (NDUFB8 and NDUFA9) in patient samples compared with controls. (C) One-dimensional blue native polyacrylamide gel electrophoresis (PAGE) (4–16% gradient) analysis showing a defect in the assembly of Complex I in patients with the homozygous NDUFB3 variant. Individual OXPHOS complexes were detected by immunoblotting using subunit-specific antibodies (Complex I (NDUFB8), Complex II (SDHA), Complex III (UQCRC2), Complex IV (COX1) and Complex V (ATP5A)). The assembly of Complexes II–V was normal in all three patient samples when compared with age-matched controls. The lower panel suggests a presence of additional, partially assembled Complex I intermediates in both control and patient samples; the upper band (indicated by *) is likely to represent the ∼650 kDa Iβ subcomplex of the hydrophobic membrane arm while the lower band (indicated by **) represents partially assembled intermediates which are only visible in patient samples. These were detected by probing with an antibody raised against NDUFB8 and are in agreement with published studies.17 In (B) and (C), SDHA (Complex II) was used as loading control.