Figure 5.
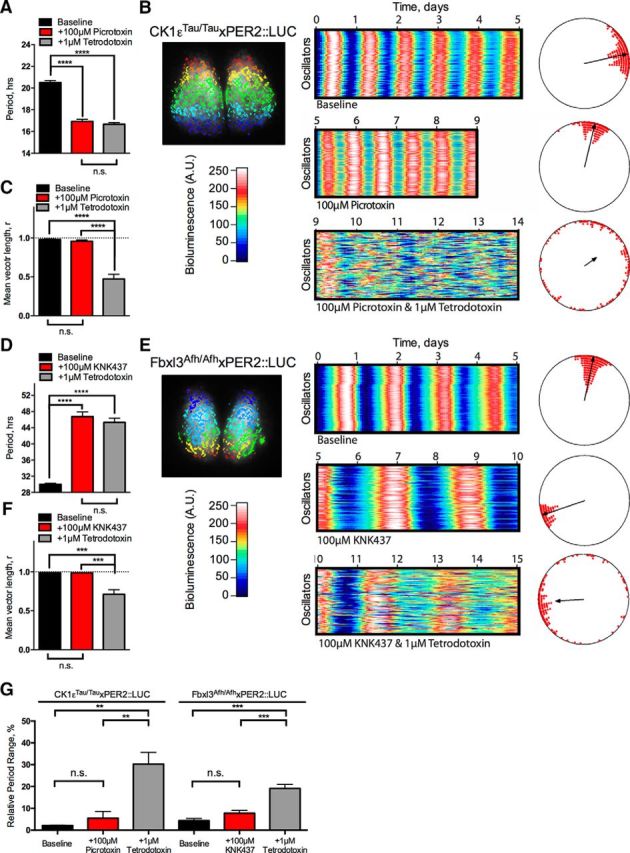
Network and cell-autonomous properties of the SCN are unaffected by pushing period to short or long extremes. A–F, SARFIA single-cell imaging analysis of CK1εTau/Tau × PER2::LUC treated with 100 μm picrotoxin (A–C) and Fbxl3Afh/Afh × PER2::LUC treated with 100 μm KNK437 (D–F). A, Summary period data from individual oscillators shown as mean ± SEM for baseline (black), 100 μm picrotoxin (+100 μm picrotoxin; red), and 100 μm picrotoxin/1 μm TTX cotreatment (+1 μm TTX; gray). B, Individual oscillators were assessed by SARFIA identification and analysis of ROIs, indicated as color-coded regions on the inset bioluminescent SCN image (left). Raster plots of 100 individual representative oscillators within the SCN are shown for baseline (top), 100 μm picrotoxin (middle), and 100 μm picrotoxin/1 μm TTX cotreatment (bottom). Relative bioluminescence intensity is color coded according to the color bar. Rayleigh plots are shown next to their corresponding raster plot. Phases of individual oscillators are plotted as circles, and the degree of synchrony is indicated by the length of the vector in the center. C, Summary synchrony data reported as mean vector length from individual Rayleigh analyses. Values are shown as mean ± SEM for baseline (black), 100 μm picrotoxin (red), and 100 μm picrotoxin/1 μm TTX cotreatment (gray). D, Summary period data from individual oscillators shown as mean ± SEM for baseline (black), 100 μm KNK437 (+100 μm KNK437; red), and 100 μm KNK437/1 μm TTX cotreatment (+1 μm TTX; gray). E, Individual oscillators were assessed by SARFIA identification and analysis of ROIs, indicated as color-coded regions on the inset bioluminescent SCN image (left). Raster plots of 100 individual representative oscillators within the SCN are shown for baseline (top), 100 μm picrotoxin (middle), and 100 μm KNK437/1 μm TTX cotreatment (bottom). Relative bioluminescence intensity is color coded according to the color bar. Rayleigh plots shown next to their corresponding raster plot. Phases of individual oscillators are plotted as circles, and the degree of synchrony is indicated by the length of the vector in the center. F, Summary synchrony data reported by mean vector length from individual Rayleigh analyses. Values are shown as mean ± SEM for baseline (black), 100 μm KNK437 (red), and 100 μm KNK437/1 μm TTX cotreatment (gray). G, Relative period range width from individual oscillators identified by SARFIA analysis in A and D expressed as a percentage of the overall cellular period. Values are shown as mean ± SEM for baseline (black), treatment (red), and TTX cotreatment (gray). Period-altering treatment conditions are detailed below the bars (+100 μm picrotoxin or +100 μm KNK437), and genotypes are detailed above the bars (CK1εTau/Tau × PER2::LUC or Fbxl3Afh/Afh × PER2::LUC). n values are detailed throughout the text. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
