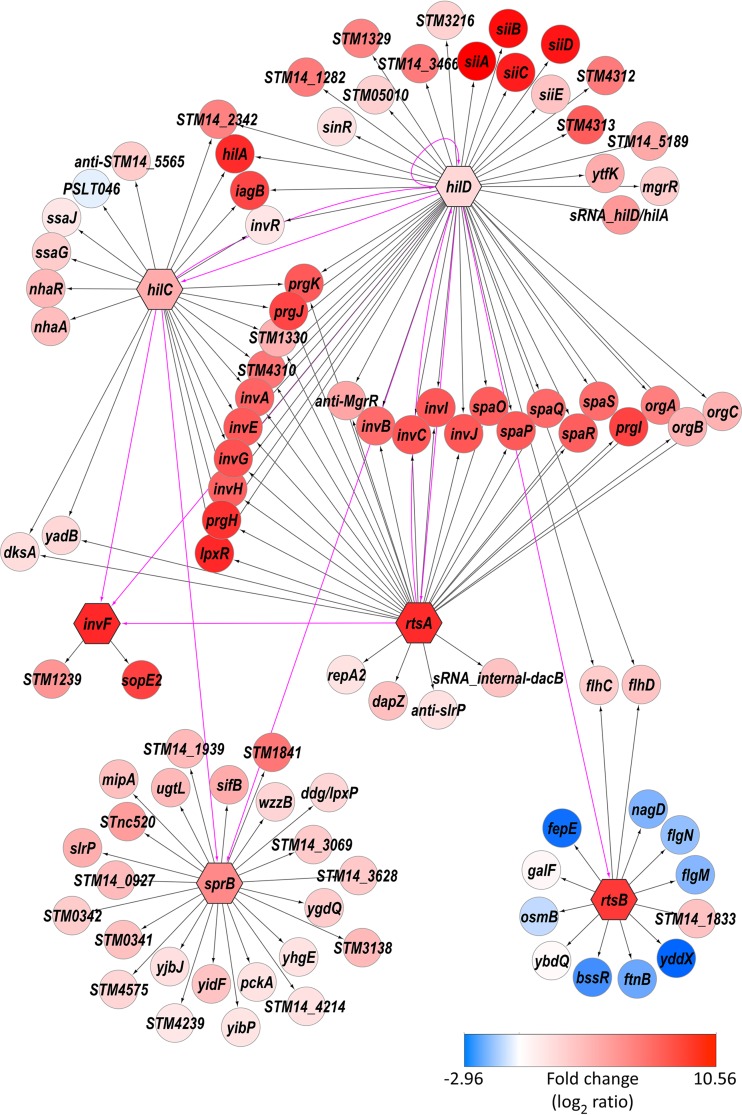
FIG 2 .
Regulatory network associated with SPI-1. The hexagon-shaped nodes represent TF-encoding genes hilD, hilC, rtsA, invF, sprB, and rtsB. The circular nodes represent target genes directly regulated by these TFs. The directed edges indicated with pink lines represent the regulatory relationships among the TFs. The directed edges indicated with black lines represent the regulatory relationships between the TFs and their target genes. The node color indicates the log ratio of the differential levels of expression (red = TF activated; blue = TF repressed), and the color scale is shown. Note that the regulatory targets of HilA are omitted because ChIP-seq for this protein was unsuccessful. Where possible, common gene names are shown. For genes without a common name, the name of the orthologous gene from S. Typhimurium strain LT2 is shown (“STMxxxx” names). For annotated genes without a common name and without an orthologue in S. Typhimurium strain LT2, the S. Typhimurium 14028s gene name is shown (“STM14_xxxx” names). For unannotated genes, we list a brief description of the transcript location (“anti” = antisense to the indicated gene; “sRNA_internal” = initiation within the indicated gene, in the sense orientation relative to that gene; “sRNA_hilD/hilA” = initiation in the intergenic region between hilD and hilA, antisense to the hilA 5′ UTR).