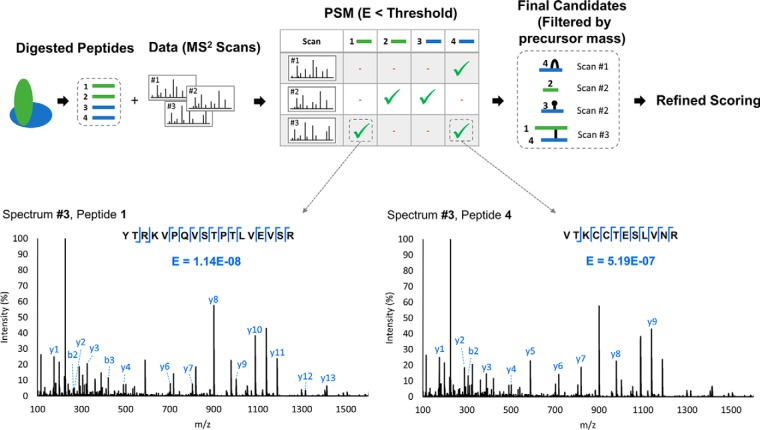
Fig. 2.
Crosslinked peptide candidate generation step involving an MS2-based library reduction strategy. A search of the full set of possible linear peptides is conducted against each MS2 spectrum using OMSSA (38), to generate a set of peptide-spectrum matches (PSM's) for each MS2 spectrum. Any peptides with an acceptable E-score for a given spectrum are allowed to form crosslinked candidates. Two PSM's are shown for Spectrum #3 for illustration: peptide 1 and 4 are strongly represented by fragment ions from the unmodified portions of peptide and are both accepted. Once the PSM's are scored and selected, all possible candidates are generated and filtered by precursor mass. Hits can include peptides that are self-linked, crosslinked, dead-ended, or simply exist as linear peptides. Candidates undergo a refined scoring method based on a rigorous peak assignment of MS and MS2 data, and a wider set of fragment ion types.