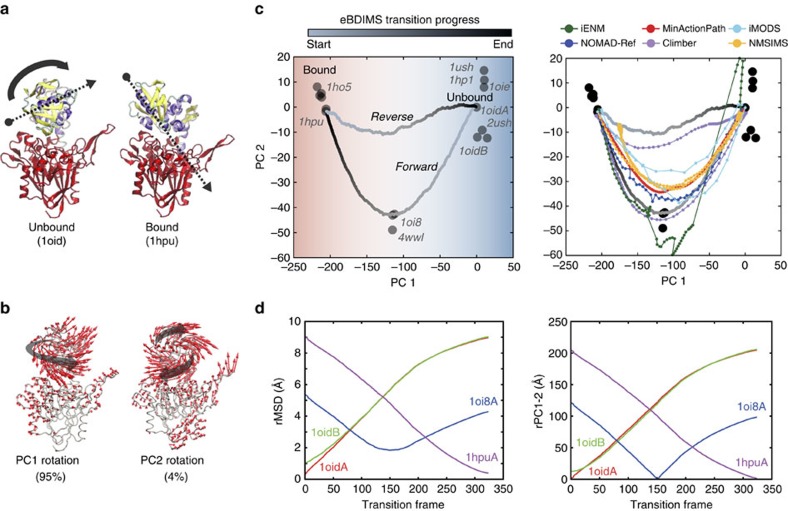
Figure 3. Conformational transition of E. coli 5′-nucleotidase (5′NTase).
(a) Crystallographic unbound (1oid) and nucleotide-bound (1hpu) states, showing one domain in red and the other coloured by secondary structure to visualize the ball-and-socket rotation (grey arrows). (b) Principal components of the X-ray ensemble (16 structures) decompose the complex rotation into a major and a minor rotation versus the reference 1oid. (c) Left: projections of the X-ray ensemble structures and the eBDIMS trajectories onto the PC1-2 subspace; PC1 alone separates the crystal structures into three clusters (same colour-coding as in Fig. 2). Right: comparison between the forward pathways computed by eBDIMS, iENM, NOMAD-Ref, MinActionPath and Climber. Reverse pathways generated by eBDIMS, iMODS, NMSIM and Climber also shown. (d) rMSD and PC1-2 distance between the forward trajectory and the sequence of crystallographic intermediates.