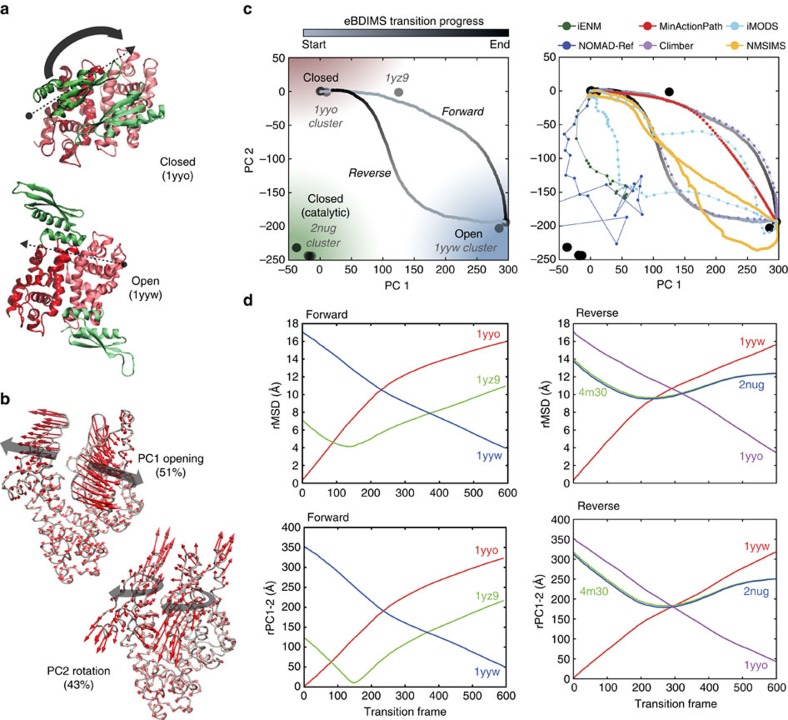
Figure 4. Conformational transition of A. aeolicus RNA endonuclease III (RNaseIII).
(a) Crystallographic dimers for closed non-catalytic state (1yyo) and open pre-catalytic state (1yyw), showing the dsRNA-binding domains (RBDs) in green and the rest of the protein in red surface; both structures are bound to dsRNA and represent different stages in re-orienting the substrate for catalysis. (b) Dominant PCs of the X-ray ensemble (11 structures) track RBD arms separation and rotation versus the reference 1yyo structure. (c) Left: projections of the ensemble and the eBDIMS trajectories onto the PC1-2 subspace reveal an hysteresis-like cycle with a tight sequence of concerted motions; PC1-2 separate the crystal structures into four clusters (shown as red (closed non-catalytic), white (intermediate), blue (open pre-catalytic) and green (closed catalytic) regions). Right: comparison between the forward pathways computed by eBDIMS, iENM, NOMAD-Ref, MinActionPath and Climber. Reverse pathways generated by eBDIMS, iMODS, NMSIM and Climber also shown. (d) rMSD and PC1-2 distance between the forward (left) and reverse (right) trajectories and the crystallographic intermediates approached.