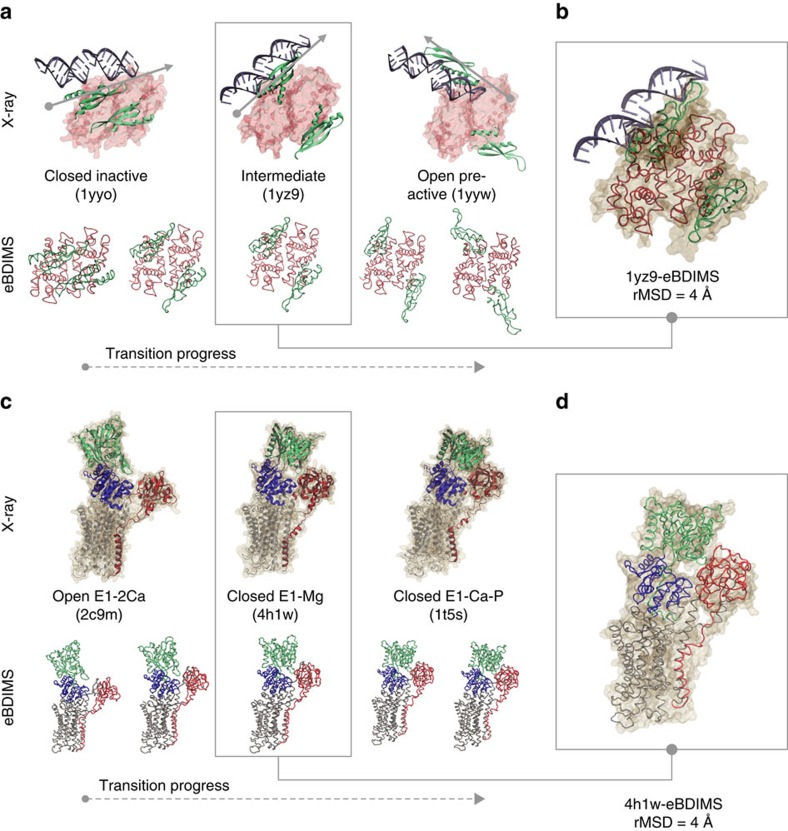
Figure 6. Comparison between the crystallographic and the eBDIMS pathways for RNaseIII and SERCA.
(a) Upper row: The complete X-ray structures for the RNaseIII end-states and the intermediate are shown bound to the dsRNA substrate; these conformations represent three sequential snapshots along the RNaseIII cycle. The starting structure is bound but catalytically inactive, and the following two show the progress to reorient the dsRNA onto the catalytic cleft (approximately in the z axis of symmetry). Lower row: as the two dsRNA-binding domains start to open the eBDIMS pathway shows how they spontaneously sample PC1 approaching the 1yz9 intermediate (shadowed); then, they leave it behind as rotation along PC2 progresses. (b) Superimposition between X-ray intermediate structure 1yz9 and the best overlapped eBDIMS frame. (c) Upper row: The complete X-ray structures for the SERCA open and closed end-states and one of the Mg2+-bound visited intermediates (4h1w) are shown; the structure is approached within 4 Å (d) by the forward eBDIMS pathway tracking headpiece closing.