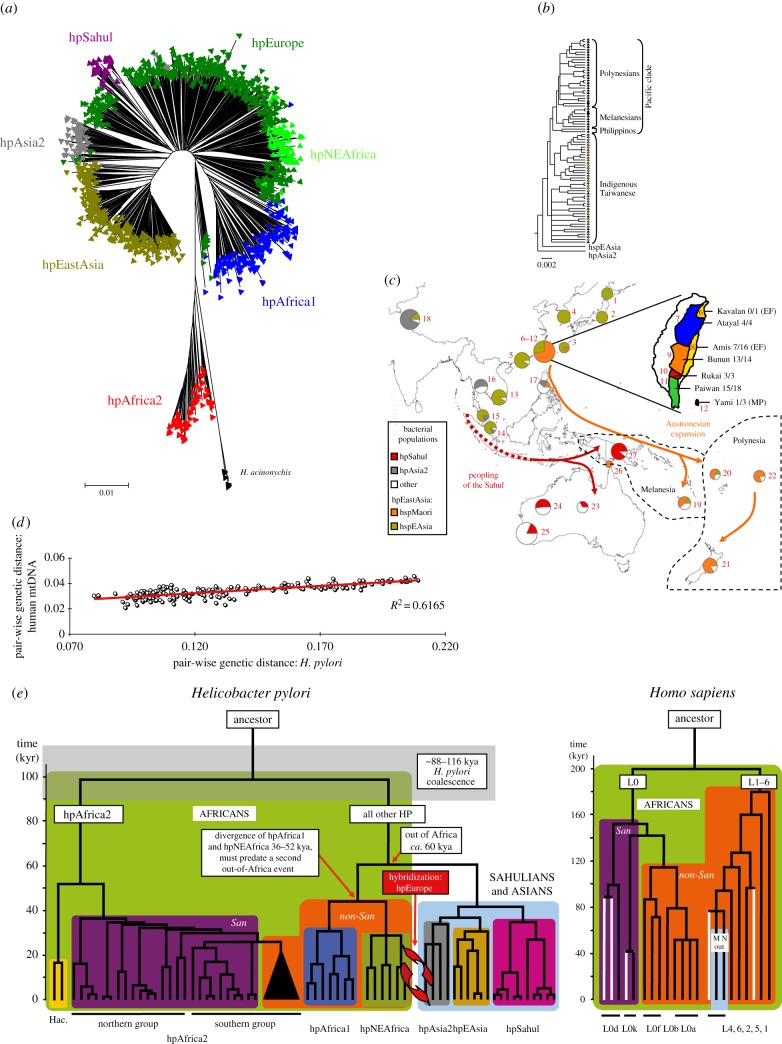
Figure 1.
Population structure, dated phylogeny and ancient migrations of Helicobacter pylori based on sequences of seven housekeeping gene fragments. (a) Global population structure (based on electronic supplementary material, figure S3 [25] updated with additional data from [22]). (b,c) Migrations into the Pacific of hspMaori and hpSahul (modified from figure 1 of [22]). (b) A phylogenetic tree of hspMaori, a subpopulation of hpEastAsia, shows patterns of serial descent from bacteria from indigenous Taiwanese of six ethnic groupings through isolates from Filipinos, Melanesians and Polynesians. (c) Frequencies of hspMaori (orange) and hpSahul (red) in pie charts according to geographical source. (c inset; Taiwan subdivided by indigenous ethnic group; number of hspMaori/number of all H. pylori (Austronesian language family)). (d) Quantitative parallel patterns of pairwise genetic distances between H. pylori and human mtDNA samples from corresponding geographical areas. (source: figure 7 of [20]). (e) Comparison of phylogenetic tree of H. pylori housekeeping gene fragments (left) and human mtDNA (right) (source: figure 6 of [20]). African lineages are shown on a green background, whereas the background for lineages outside Africa is light blue. San clades are purple, non-San clades are orange and H. acinonychis is yellow. San mtDNA lineages that were identified in this study are shown as white lines.