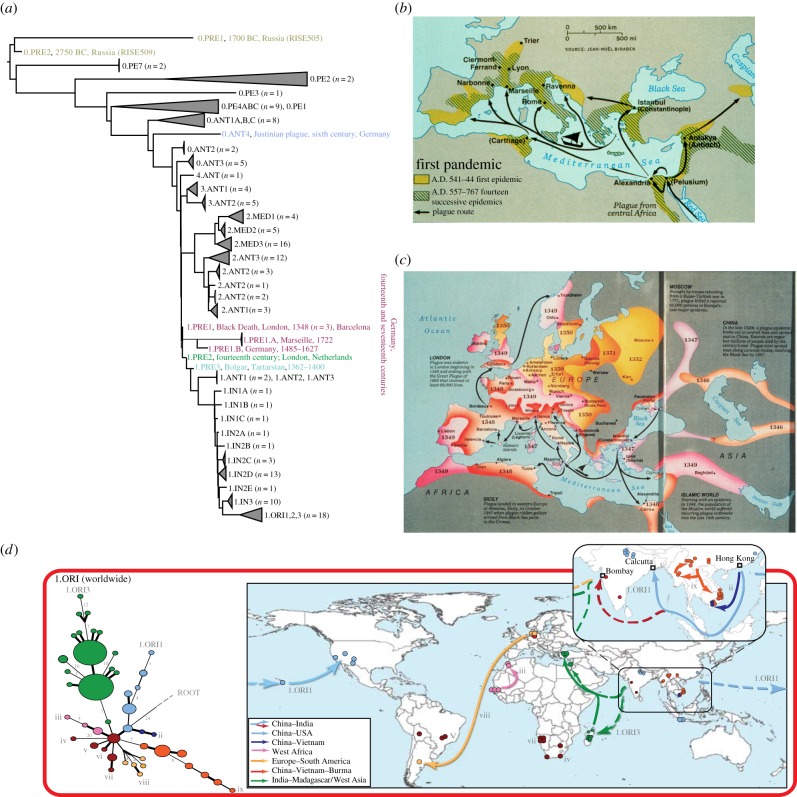
Figure 2.
Plague pandemics and phylogeny of Yersinia pestis. (a) Manual concatenation of the topologies of phylogenies described in [30], [16] and [31], including additional ancient genomes and genotypes (see below), plus proposed mnemonics for those populations. Sources of ancient genomes and genotypes: the Bronze Age (0.PRE1, 0.PRE2; grey) [31], the Justinianic Pandemic (0.ANT4; blue) [32]; the Black Death (1.PRE1; maroon; London, 1348 [33]; Barcelona 1300–1420 [34]; SNP-based genotypes from Germany (fourteenth and seventeenth centuries) [35]) and descendent branches of 1.PRE from Ellwangen (1.PRE1B; 1485–1627) [34] and Marseille (1.PRE1.A, 1722) [36]). One SNP further along branch 1 are the populations 1.PRE2 (London, 1362–1400 [36] and the related low-resolution genotype found in Berg-Op-Zoom, The Netherlands from the fourteenth century [37]; green). One further SNP down the branch is a short branch leading to a genome from Western Asia (1.PRE3; Bolgar City, Tatarstan, Russia; light blue) [34]. (b,c) Maps of the spread of the Justinianic (b) and second plague (c) pandemics. With permission from Elisabeth Carniel. (d) Reconstruction of waves of transmission of individual lineages within the third pandemic. Modified from supplementary figure 3 from [30] (http://research.ucc.ie/NG1/index.html).