Figure 3.
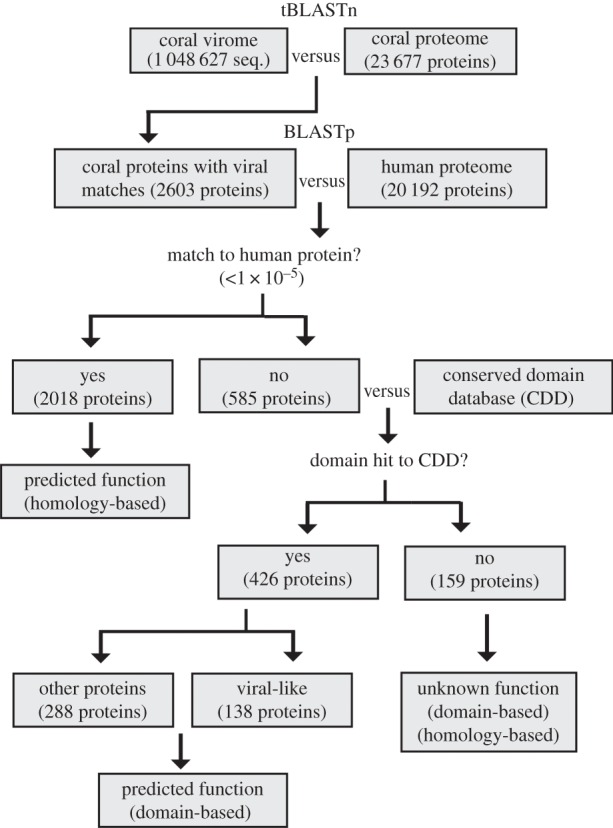
Bioinformatic pipeline used to identify viral gene segments with sequence similarity to host proteins: uncharacterized viruses versus uncharacterized immune system (e.g. corals). Coral viromes extracted from four species of coral were compared with the predicted Acropora digitifera proteome through tBLASTn analysis resulting in 2503 protein hits. Coral proteins were classified as ‘hits’ if five or more unique viral sequences matched with e-value less than 1 × 10−4 (see electronic supplementary material, table S3 for a summary of viral hits to coral proteins and electronic supplementary material, file S2 for full tBLASTn results). To determine whether the function of the 2503 coral proteins could be predicted based on sequence similarity to human proteins they were compared with the human proteome using BLASTp with an e-value cutoff of 1 × 10−5 resulting in 2018 coral proteins with positive matches to human proteins (see electronic supplementary material, file S3 for full BLASTp results). To determine whether any of the remaining 585 proteins function could be predicted based on the presence of conserved domains they were analysed using the conserved domain database (CDD) resulting in 426 coral proteins with positive matches to a known domain (see electronic supplementary material, file S4 for full results of CDD analysis). Within this group of proteins 138 were considered ‘viral-like’ based on the presence of domains associated with viral proteins. In total 159 coral proteins possessed no similarity to either database and therefore represent predicted immune proteins with unknown function.
