Figure 5.
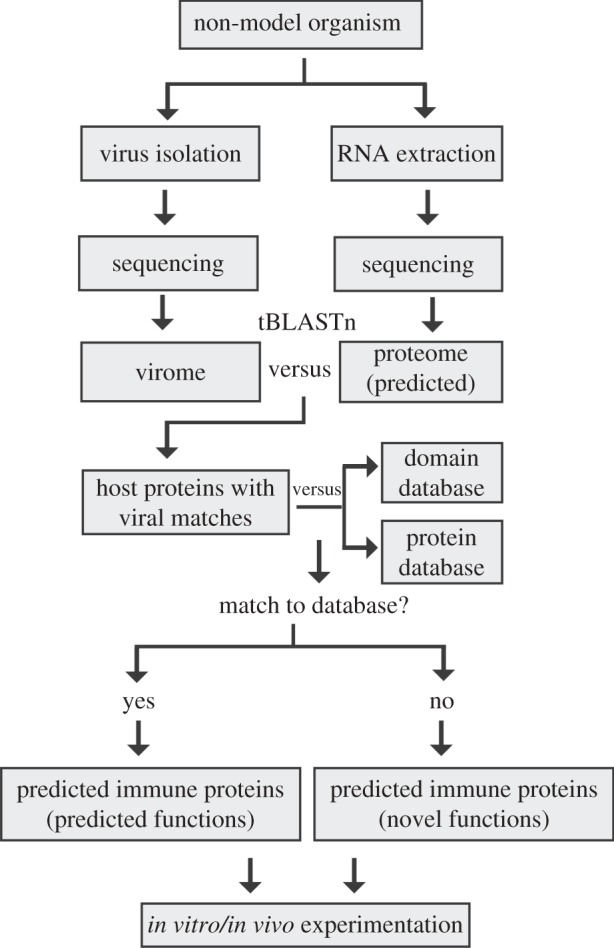
Proposed pipeline to predict immune proteins in uncharacterized systems. Viruses are extracted from the non-model organism of interest using a virus-like particle extraction protocol of choice and nucleic acid is sequenced. If a gene model is not available total RNA is also extracted from host tissue and an assembled transcriptome is prepared. Viral gene segments are compared with the translated transcriptome through tBLASTn to identify matches to host proteins. These proteins are further analysed through comparison with a well-characterized immune system using BLASTp (e.g. human or mouse) and domain prediction database (e.g. conserved domain CDD, Pfam). Proteins that lack hits to either database represent predicted immune genes with unknown function and are selected for further biochemical investigations using in vitro and in vivo experimentation.
